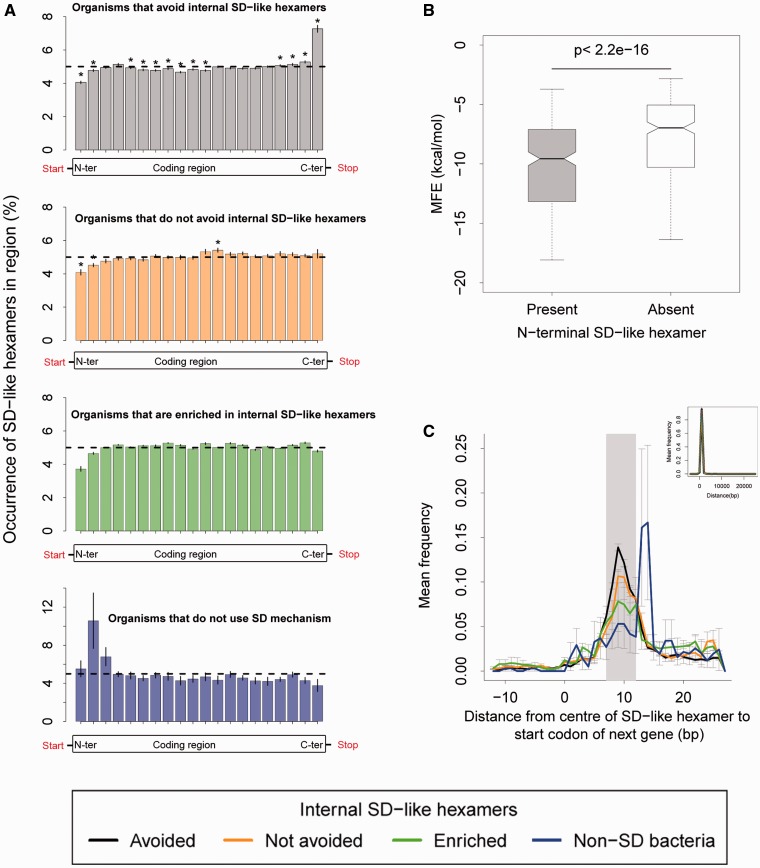
Fig. 5.—
Functional and positional analysis of the occurrence of internal SD-like hexamers (aff5–highest affinity). (A) The distribution of SD-like hexamer occurrence (mean ± SEM) across the relative length of all genes in 284 organisms, for genomes that avoid, do not avoid, are enriched in internal SD-like hexamers and organisms that do not use the SD mechanism of translation initiation. Dashed lines indicate the random expectation for occurrence in each region (gene length/20 regions = 5%). In each panel, asterisks indicate gene regions where the occurrence was significantly different from this expectation (one-sample Wilcoxon signed rank test with Benjamini–Hochberg correction for multiple comparisons, P < 0.05) (B) N-terminal MFE of genes with an N-terminal SD-like hexamer (grey bars) versus those without (white bars) for 277 organisms that use the SD mechanism of translation initiation. The two distributions were significantly different from each other (paired Wilcoxon test, P < 2.2e−16). (C) The mean frequency distribution (±SEM) of the distance from the centre of a C-terminal SD-like hexamer to the start codon of the next gene (inset plot shows the entire distribution). The grey rectangle indicates the most common distances between the ribosome binding site and the start codon of genes in E. coli (7–12 bp).