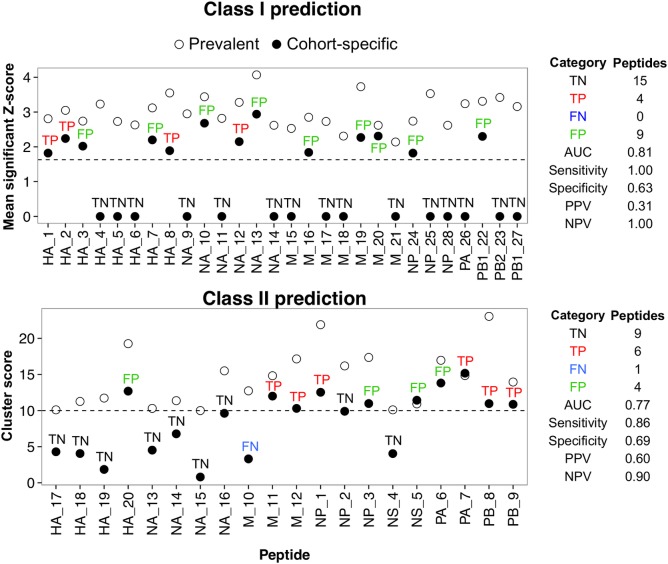
Fig 5. Comparison between prediction for prevalent and cohort-specific SLA alleles.
Peptides were predicted to bind to a set of previously reported class I and class II SLA alleles prevalent in the U.S. swine population (prevalent). Based on low-resolution SLA-typing results, those alleles were not represented in the studied pigs. Prediction matrices were developed to predict binding potential of peptides to the most frequent SLA alleles found in the cohort (cohort-specific). (Top) Mean of significant Z-scores (above 1.64) over prevalent class I SLA alleles (SLA-1*0101, 1*0401, 2*0101, and 2*0401) and cohort-specific (SLA-1*0801, 1*1201, 1*1301, 2*0501, and 2*1201) are shown for each peptide. Peptides with a mean of significant Z-scores above 1.64 (dashed line) are considered potential binders. (Bottom) Cluster scores calculated for prevalent class II SLA alleles (DRB1*0101, 0201, 0401, and 0601) and cohort-specific alleles (DRB1*0402, 0602, 0701, and 1001) are shown for each peptide. Cluster scores above 10 (dashed line) are considered as potential binders. Based on the retrospective evaluation, peptides were classified in four categories (TN: true negatives, TP: true positives, FN: false negatives, and FP: false positives). AUC, Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) are shown.