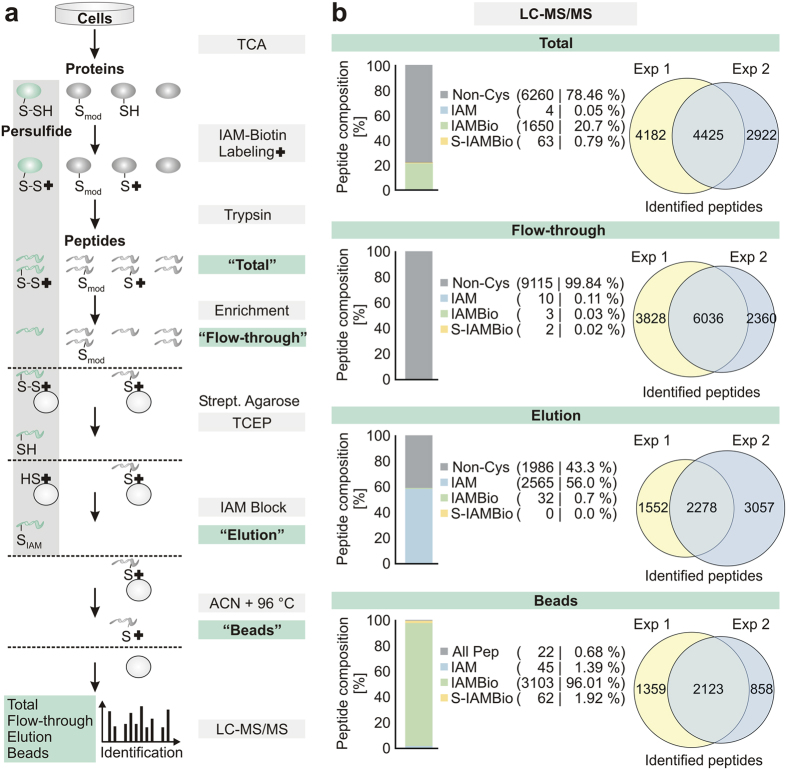
Figure 1. Workflow for the identification of persulfide containing peptides using mass spectrometry.
(a) Cells were subjected to trichloroacetic acid (TCA) precipitation and subsequently, the thiols and persulfides were labelled using iodoacetyl-PEG2-Biotin (IAMBio). After digestion of the proteins (Total), single peptides containing either labelled persulfides or cysteines were enriched and separated from non-cysteine peptides (flow-through) using streptavidin agarose beads. After several washing steps, persulfide containing peptides were eluted using tris(2-carboxyethyl)phosphine (TCEP) where thiol containing peptides were not affected. The subsequent accessible cysteines were labelled with iodoacetamide (IAM) (Elution). As control, labelled thiol peptides remaining on the beads were eluted using 10 mM TCEP and 80% acetonitrile (ACN) (Beads). Samples of the total, flow-through, elution and bead fraction were subjected to liquid chromatography and mass spectrometry (LC-MS/MS) and the peptides were identified using the PEAKS 7.0 proteomics software. (b) Number of identified peptides in the different fractions as described in a). Shown are the mean values of two independent experiments. S-IAMBio: Persulfide peptide modified by IAMBio.