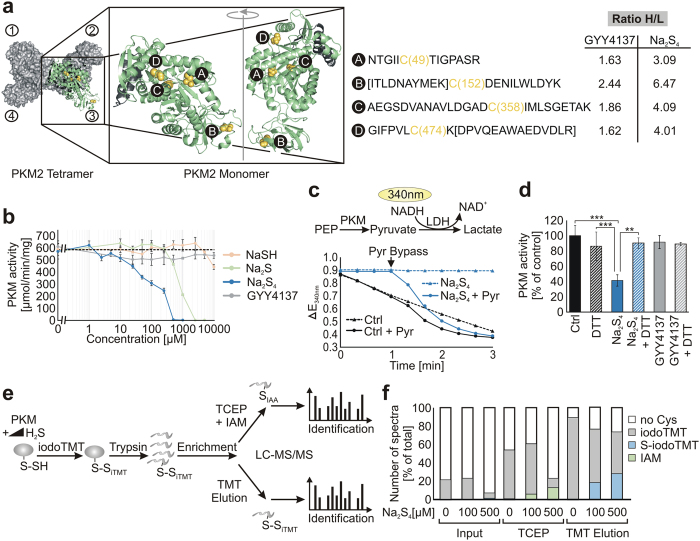
Figure 5. In vitro characterisation of persulfide formation on PKM2.
(a) Structure showing the human PKM2 monomer and tetramer (PDB: 3SRH). Cysteine peptides forming persulfides are highlighted (yellow spheres; A, B, C, D). Black: intersubunit contact site; letters in square brackets: sequence of misscleaved peptide. (b) Purified PKM2 from rabbit muscle was incubated with increasing concentrations of Na2S4, Na2S, NaSH and GYY4137. PKM2 activity (μmol/min/mg) was measured in a coupled enzyme assay with LDH as second enzyme monitoring the consumption of NADH at 340 nm. (c) The activity of PKM2 was followed as decline in absorption at 340 nm. After 1 min 5 mM pyruvate was added to bypass the reaction catalysed by PKM2. (d) The experiment was carried out as described in (b) except that 1 mM DTT was added in parallel to treatment with 200 μM Na2S4. Data are means +/− SD, **p < 0.01 Na2S4 vs Na2S4 + DTT, ***p < 0.001 Ctrl as well as DTT vs Na2S4. (e) Workflow to confirm that persulfides are formed at PKM2. PKM2 was incubated with 100 μM and 500 μM Na2S4 or kept untreated. Induced persulfides were modified with iodoTMT similar to qPerS-SID. After digestion with trypsin the persulfide peptides were enriched using an anti-iodoTMT resin and subjected to TCEP elution followed by IAM blocking as described for the proteomic approach. In parallel, direct labelled persulfides (S-iodoTMT) were eluted using iodoTMT elution buffer. The eluted peptides were subjected to LC-MS/MS measurement and the peptides were identified using PEAKS 7.0. (f) Spectra counts of iodoTMT labelled cysteine peptides (iodoTMT), persulfide peptides identified according to the qPerS-SID protocol (TCEP elution, IAM) and iodoTMT labelled persulfide peptides (TMT elution, S-iodoTMT).