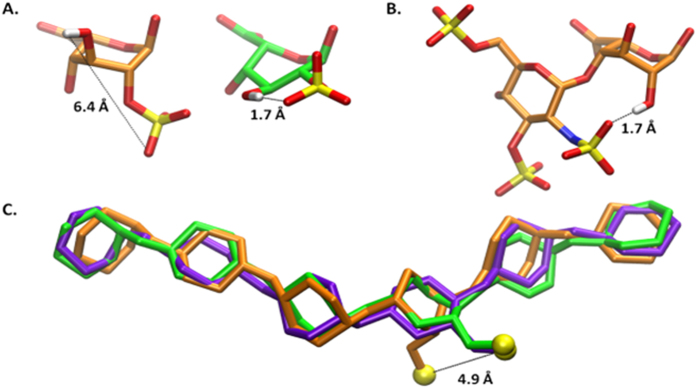
Figure 3. Images of IdoA2S or IdoA and hexasaccharides from the MD simulations.
Atomic distances are indicated by dashed lines. The 1C4, 2SO, and 4C1 conformations of IdoA2S are colored orange, green, and purple, respectively. Panel A shows the IdoA2S residue in the 1C4 and 2SO conformations (from a representative snapshot of the simulations of 4). Panel B displays the GlcNS(3S,6S) and IdoA residues from a representative frame of the simulation of 8 in the 2SO conformation. Panel C contains a superimposition of the ring atoms in oligosaccharides from simulations of 4 in each of the three IdoA ring conformations. The sulfur atom of the IdoA2S residue is also displayed in yellow for each conformation. The RMSD of the ring atoms of the ligands with IdoA in the 1C4 and 4C1 conformation are 0.9 Å and 0.7 Å, respectively, relative to the molecule containing IdoA in the 2SO conformation. Each model is the snapshot most similar to the average structure during the MD simulations.