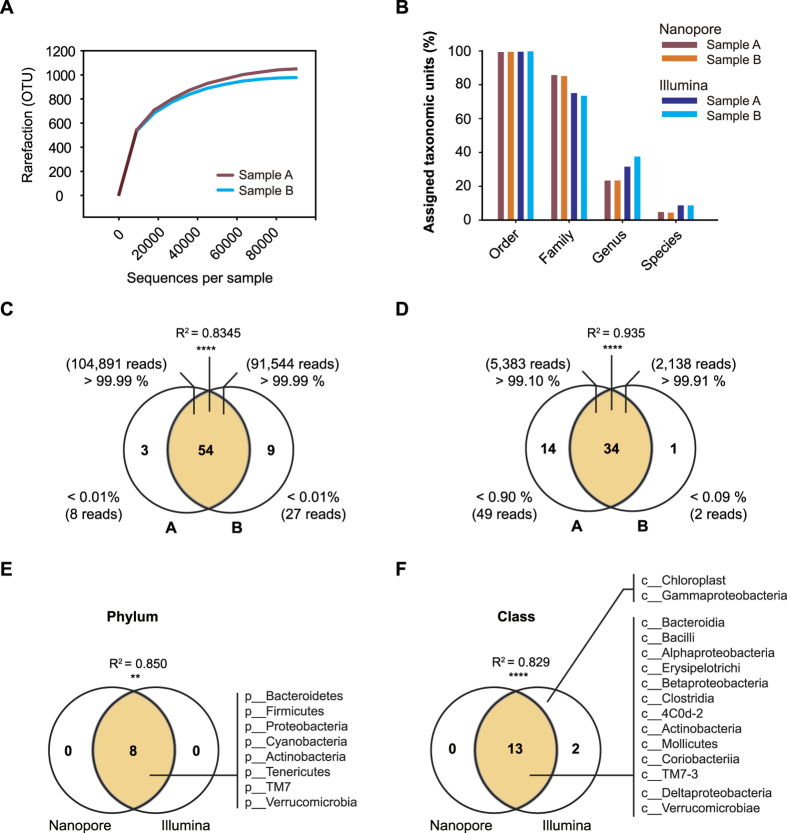
Figure 2. Statistical comparison between short-read and nanopore sequencing data.
(A) Rarefaction curves of mouse fecal samples based on short-read sequencing (Illumina). Total OTUs were generated by 3% distances. Total sample richness estimates were calculated by the observed OTUs. (B) Percentage of taxonomic units assigned as reads at the order, family, genus, and species levels. (C) Venn diagram showing the shared and specific phylotypes between Illumina A and B data. The Spearman rank correlation test (R2 = 0.8345, p < 0.0001) showed the significance of relationships between duplicates. Asterisks indicate the significance of the pairing (****p < 0.0001) (D) Two-way Venn diagram depicting the number of shared and specific phylotypes between nanopore A and B data. Percentages show the proportion of aligned reads corresponding to each phylotype per total reads. The results of Spearman rank correlation test (R2 = 0.9350, p < 0.0001) showed the significance of relationships between duplicates. Asterisks indicate the significance of the pairing (****p < 0.0001). (E,F) Venn diagram showing the shared and specific taxonomic units at the (E) phylum and (F) class levels between nanopore and Illumina sequencing data. The Spearman rank correlation test showed the significance of the relationship. Asterisks indicate the significance of the pairing (** 0.01 < p < 0.001, ****p < 0.0001).