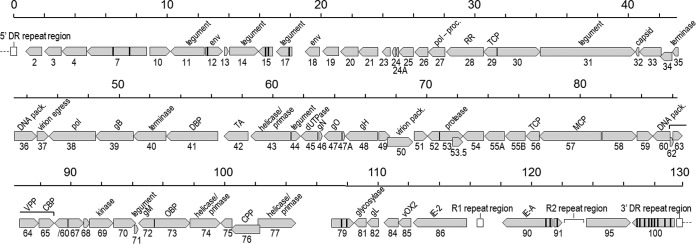
FIG 2.
Map of the ORFs identified in the MneHV7 genome. The genomic positions (130-kb unique genome sequence, excluding DRs), lengths, and transcription directions of 84 MneHV7 ORFs (in gray) were identified on the basis of similarity with the annotated HHV-7 RK strain genome using GATU software. ORF boundaries were determined on the basis of the position of start and stop codons and, when applicable, confirmed by the presence of appropriately located poly(A) signals and TATA boxes. Vertical black lines within an ORF indicate predicted splice sites, while the longer-ranged splice between the two protein-coding exons in U60 is indicated with a bar. Internal repeat regions R1 and R2 and the end-terminal direct repeat regions (DR-L [5′], DR-R [3′]) are represented by white rectangles. proc., processing; pack., packaging; RR, ribonucleotide reductase; TCP, triplex capsid protein; DBP, single-stranded DNA-binding protein; MCP, major capsid protein; CPP, capsid portal protein; TA, transactivator.