FIG 3.
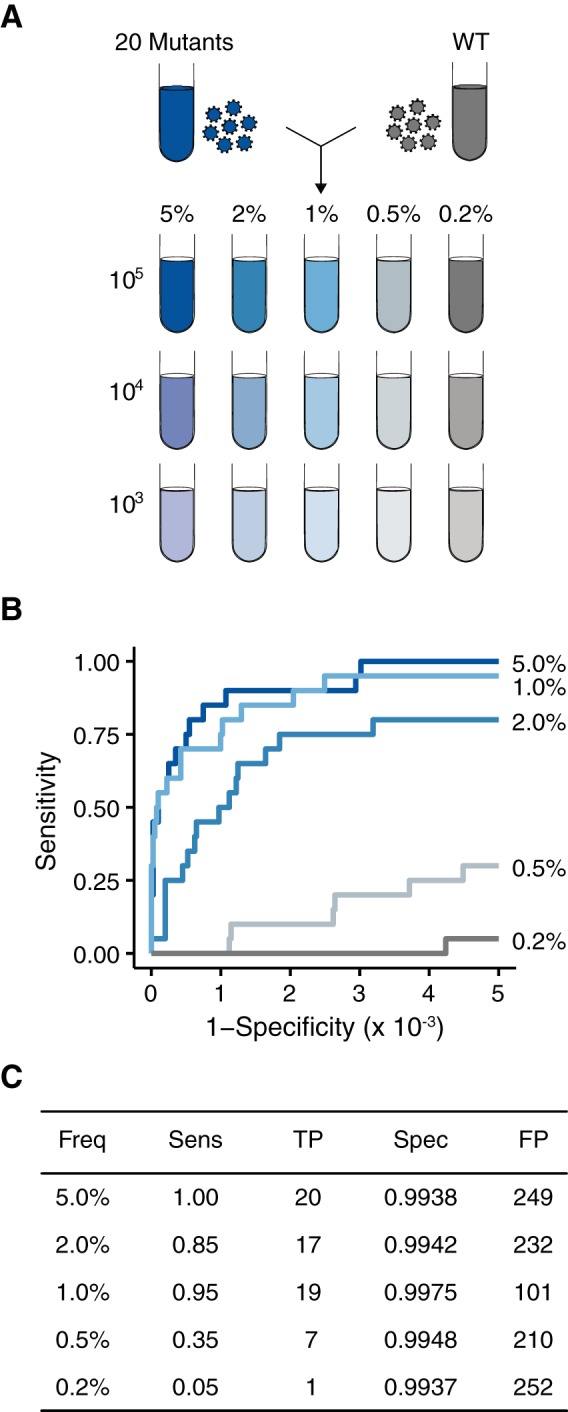
Accuracy of DeepSNV on populations approximating patient-derived samples. (A) Twenty viral supernatants, each with a single SNV, were diluted in a WSN33 viral supernatant to generate artificial viral populations with 20 mutations at the indicated frequencies. These populations were diluted further in basal medium to match the genome concentrations found in patient-derived samples (105 to 103 genomes/μl). (B) ROC curve measuring the accuracy of DeepSNV in identifying SNV at the indicated frequencies. (C) Summary of the data in panel B at a P value threshold of 0.01.
