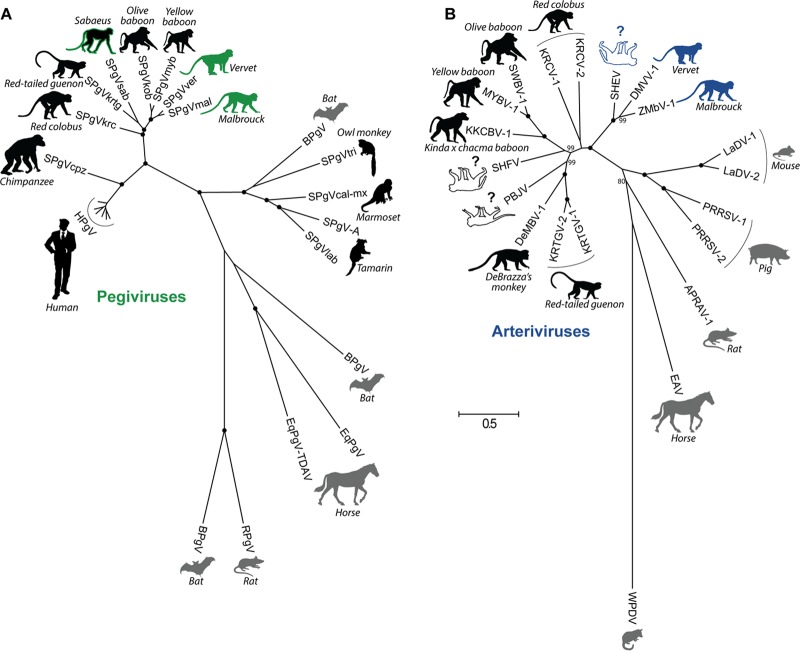
FIG 1.
Species-level phylogenetic relationships of all known pegiviruses (A) and arteriviruses (B). Viruses are shown adjacent to the silhouette of their respective host, with host common names in italics. Primate hosts are shown in black, and nonprimate hosts are shown in gray. Viruses discovered in this study are depicted by host silhouettes with solid coloring: green for AGM pegiviruses and blue for AGM arteriviruses. Host silhouettes with a colored outline draw attention to viruses of importance. Sabaeus SPgV (SPgVsab) has a green outline because this virus does not group with the AGM pegiviruses presented here. SHEV has a blue outline because of its close relationship to the AGM arteriviruses presented here. White host silhouettes symbolize arteriviruses that have caused outbreaks of viral hemorrhagic fever in captive macaques. Question marks emphasize that the natural host(s) of these viruses remains unknown. Shown is a maximum likelihood tree with 1,000 bootstrap replicates. Black dots indicate splits that are supported by 100% of bootstrap replicates. Bootstrap values below 70 are not shown. The bar shows the calculated genetic distance. SPgVkrc, Kibale red colobus; SPgVkrtg, Kibale red-tailed guenon; SPgVob, SPgV from olive baboon; SPgVmyb, Mikumi yellow baboon; BPgV, bat pegivirus; SPgVtri, owl monkey; SPgVcal-mx, marmoset-mystax; SPgVlab, tamarin; EqPgV, equine pegivirus; TDAV, Theiler's disease-associated virus; RPgV, rat PgV; KRTGV-1, Kibale red-tailed guenon virus; DeMBV-1, Debrazza's monkey virus; PBJV, Peter B. Jahrling virus; KKCBV-1, Kafue kinda-chacma baboon virus; MYBV-1, Mikumi yellow baboon virus; SWBV-1, Southwest baboon virus; KRCV-1, Kibale red colobus virus; LaDV-1, lactate dehydrogenase elevating virus; PRRSV-1, porcine reproductive and respiratory syndrome virus; APRAV-1, African pouched rat virus; EAV, equine arteritis virus; WPDV, wobbly possum disease virus.