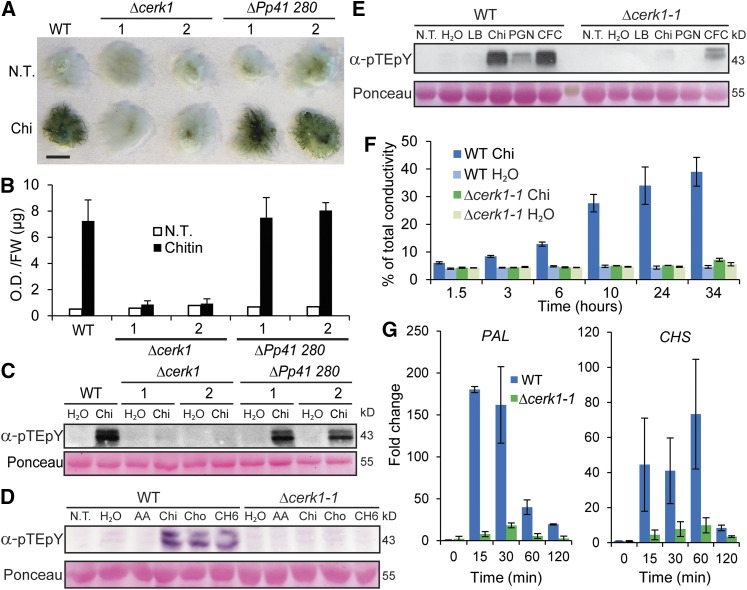
Figure 1.
Characterization of Chitin Receptor Knockout Lines.
(A) Toluidine blue staining of untreated (N.T.) and 48 h chitin (Chi) treated wild type and targeted (KOs) Pp CERK1 (Δcerk1-1 and 2) and another homolog (ΔPp41 280-1 and 2) of Arabidopsis CERK1. Bar = 5 mm.
(B) Quantitation of stainings in (A) (see Methods).
(C) Chitin (Chi) induced MAP kinase phosphorylation in the wild type and ΔPp41 280 but not in Δcerk1 KO lines assayed by immunoblotting with anti-phospho-p44/42 MPK antibody (α-pTEpY). Loading control is Ponceau-stained total protein.
(D) Immunoblot assay for MAP kinase phosphorylation of wild type and Δcerk1-1 KO line in response to no treatment (N.T.), water, 0.01% acetic acid (AA), 100 µg/mL chitin (Chi), 100 µg/mL chitosan (Cho), or 100 µg/mL hexa-N-acetylchitohexaose (CH6) for 5 min.
(E) Immunoblot assay for MAP kinase phosphorylation of wild type and Δcerk1-1 KO line in response to no treatment (N.T.), water, Luria broth (LB), 100 µg/mL chitin (Chi), 200 µg/mL peptidyl glycan from Staphylococcus aureus (PGN), or cell-free culture filtrate (CFC) from the harpin negative strain P. wasabiae SCC3193.
(F) Tissue ion leakage assay of the wild type and Δcerk1-1 treated with water or chitin for times specified. Data are presented as percent of total conductivity as an average of four replicates each with five plants. Error bars represent sd.
(G) RT-qPCR of PAL4 and CHS transcript levels in the wild type and Δcerk1-1 as fold change relative to the wild type untreated at times specified after 100 µg/mL chitin treatment. Data points represent an average of three independent experiments (three biological replicates) each with three technical replicates for each PCR reaction. Error bars represent se of the means.