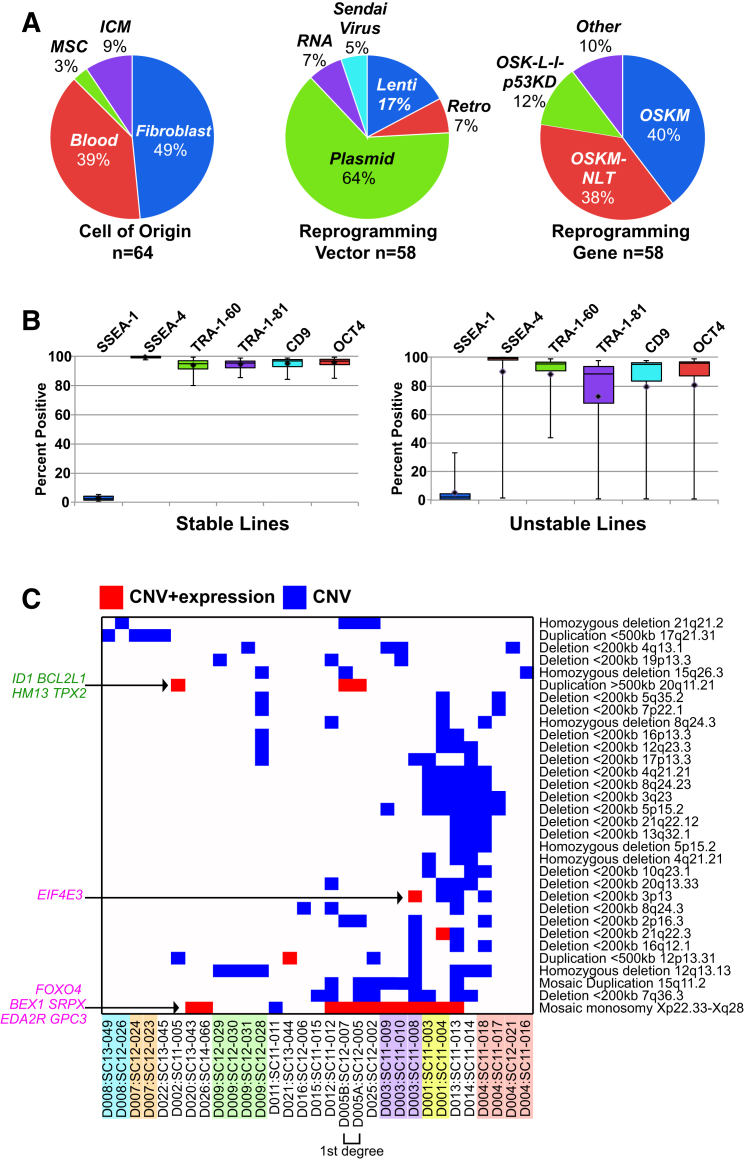
Figure 2.
iPSC Line Characteristics, Flow Cytometry Analysis, and CNV Accumulation
(A) Reprogramming variables for: originating cell type (left), reprogramming vector (middle), and gene combination (right). Reprogramming gene combinations: OSKM is composed of POU5F1 (also known as OCT4), SOX2, KLF4, and c-MYC; OSK-L-l-p53KD includes LIN28A and TP53 knockdown and l-MYC instead of c-MYC; OSKM-NLT includes NANOG, LIN28A, and SV40 large T antigen. ICM, inner cell mass; MSC, mesenchymal stem cell.
(B) Flow cytometry analysis classified iPSC as stable (n = 41) or unstable (n = 11). Boxes represent the first and third quartiles, whiskers show the complete range, and the horizontal line is the median.
(C) Commonly observed CNV predicted as non-benign or clinically significant and observed from at least three independent genetic donors are listed on the right and shown as a heatmap (blue). Red cells indicate that CNV overlaps genes with concordant expression differences. Concordant genes with known function are labeled on the left, with previously identified tumor-suppressor genes in purple and cell-growth-promoting and oncogenesis-promoting genes in green. Lines from the same donor are highlighted in the same color.