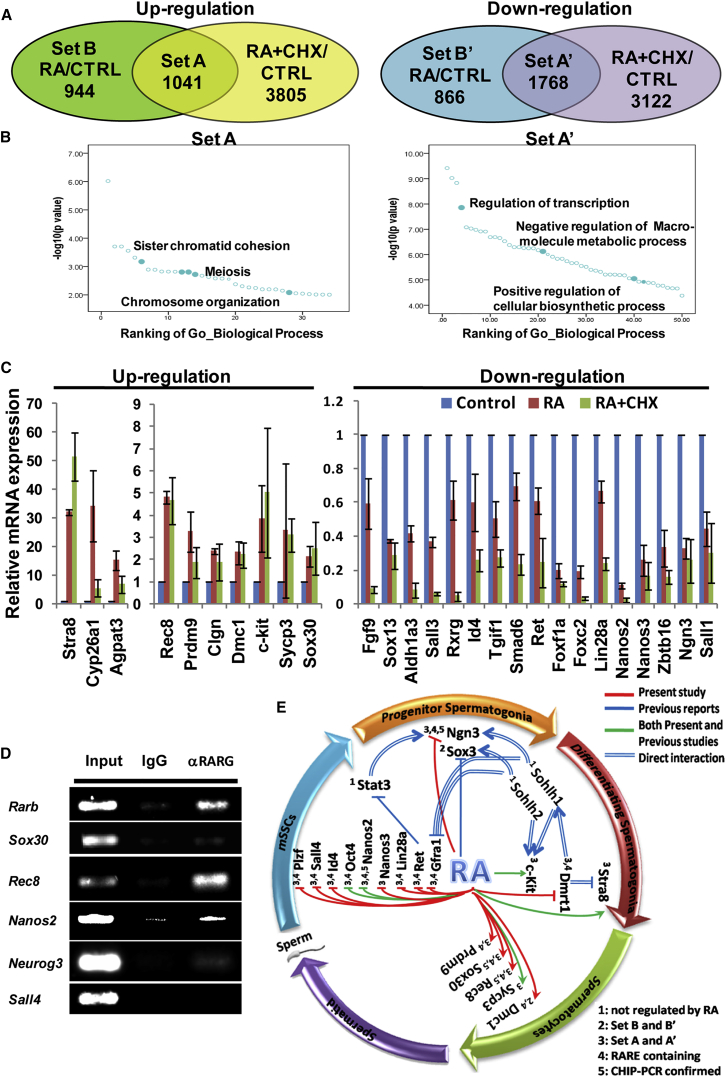
Figure 2.
A Network of Genes Regulated by RA Signaling
(A) Up- and down-regulated genes by RA and RA + CHX and their intersections. Feeder-free and serum-free mSSCs were treated with vehicle or RA or RA plus cycloheximide (RA + CHX) for 24 hr, and then total RNA from different samples was extracted for RNA-seq analysis.
(B) Gene ontology term analysis of RA-regulated genes. Left and right panels show the significantly enriched terms for up- and down-regulated genes, respectively.
(C) qRT-PCR confirmation of a panel of RA-regulated genes. Most of these genes are well known for their roles in spermatogenesis. Error bars denote mean ± SD from three independent assays.
(D) Identification of the several potential direct target genes of RA/RAR by ChIP-PCR. Putative RARE in the promoters of Rarb, Sox30, Rec8, Nanos2, and Neurog3 were identified by motif scanning using a bioinformatics method. ChIP assays were conducted using the RARG antibody.
(E) A small network of genes regulated by RA was constructed manually based on our RNA-seq data and literature mining. Genes reported to be involved in mSSC self-renewal, progenitor spermatogonia proliferation, differentiation and meiosis initiation of differentiating spermatogonia, and meiosis of spermatocytes were placed underneath the corresponding stages of spermatogenesis. Regulatory interactions identified by the present study (red line) or previous reports (blue) or both (green) were labeled with lines of different color. The double lines indicate a direct binding of a transcription factor to the promoters of their target genes. The numbers denote different types of evidence for the regulations by RA. 1, 2, 3 indicate RNA-seq data; 4, bioinformatics scanning of the RARE; and 5, ChIP-PCR results.
See also Figures S2 and S3.