Figure 6.
Transcriptome Profiles of Induced Germ Cells and Isolated Spermatogenic Cells
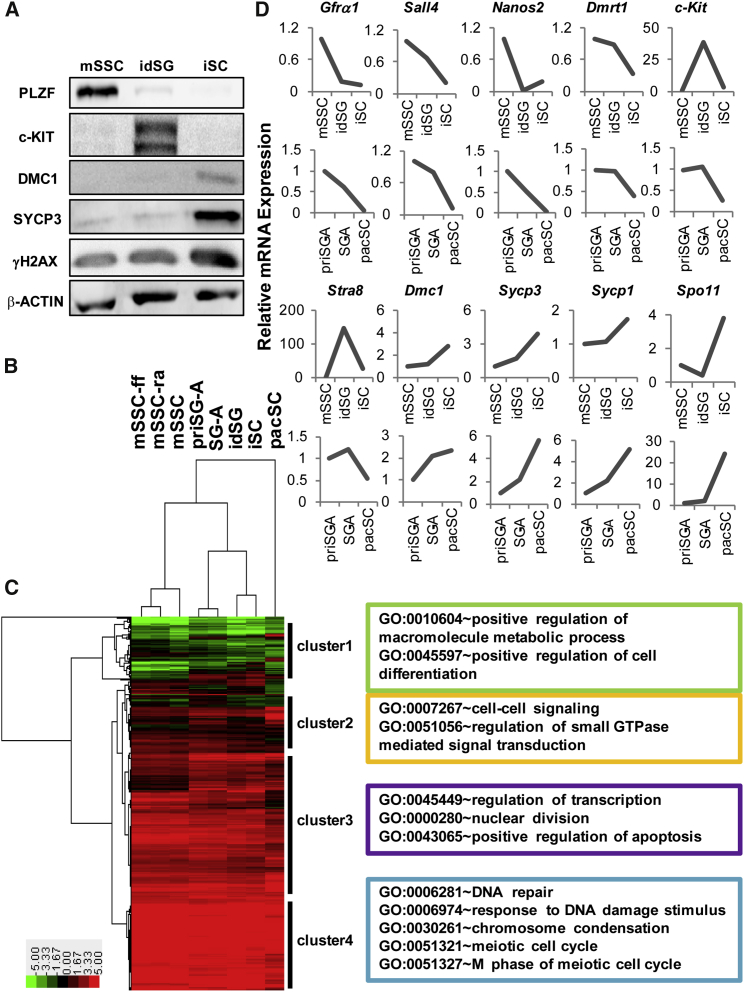
(A) Characterization of three types of in vitro spermatogenic cells by Western blotting analysis of several marker proteins specific to different spermatogenic cells. idSG, induced differentiating spermatogonia; iSC, induced spermatocytes.
(B and C) Hierarchical clustering of in vitro- and in vivo-derived spermatogenic cells (B) and protein-coding genes (C) based on the RNA-seq data. mSSC-ff, feeder-free mSSCs; mSSC-ra, RA-treated mSSCs; priSG-A, primitive SG-A isolated from 6-dpp mice; SG-A, isolated type A spermatogonia; pacSC, isolated pachytene spermatocytes. Differentially expressed genes were identified using ANOVA and samples of mSSC-ff, mSSC-ra, mSSC, idSG, and iSC, each of which included two biological replicates. Four clusters of genes were labeled based on visual inspection of the hierarchical clustering of genes, and representative enriched gene ontology (GO) terms are listed on the right side of the heatmap.
(D) Comparison of the expression patterns of a set of genes involved in spermatogenesis in the in vitro- and in vivo-derived germ cells using the FPKM values (fragments per kilobase of transcript per million mapped reads) of the RNA-seq data.
See also Figure S7.