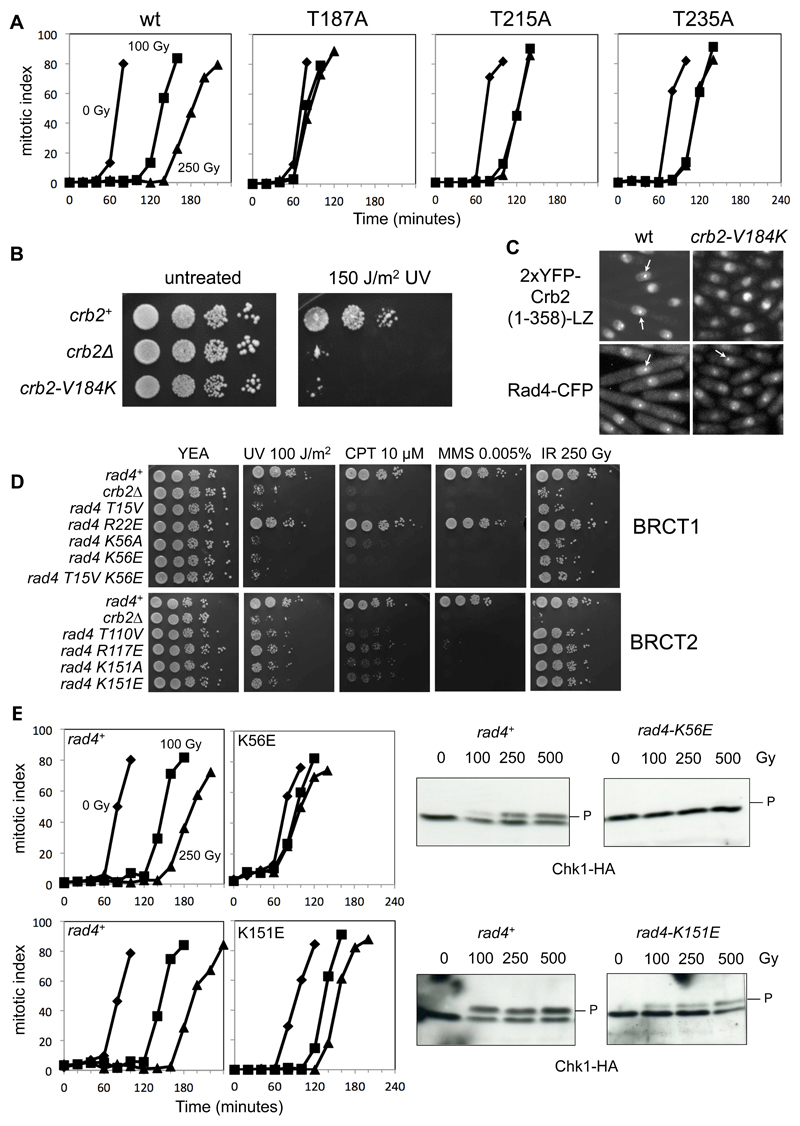
Figure 5. Rad4-Crb2 phospho-dependent interactions are essential for a DNA-damage checkpoint response.
A) Cell cycle arrest due to DNA damage, in wild-type cells and those with mutations in Thr187, Thr215 and Thr235. While wild-type cells experience a delay of 60 minutes in the time for 50% of the cells to go through mitosis, this is greatly reduced in the phosphorylation site mutants. As with survival of DNA damage, the Thr187 mutant is the most severely affected.
B) Val184 binds to the hydrophobic pockets of both Rad4-BRCT1 and BRCT2 in the crystal structure of the complex with the Crb2-pThr187 peptide. Mutation to a residue incompatible with that interaction (V184K), disrupts survival of DNA damage to a similar degree as the Crb2 null strain.
C) Crb2-V184K mutation disrupts co-localisation of Rad4 and Crb2 at DNA damage induced foci.
D) Mutation of residues in the phosphate-binding sites of either BRCT1 or BRCT2 domains of Rad4 significantly impair survival of a range of genotoxic insults. Unlike the topologically equivalent Arg117 in BRCT2, Arg22 in BRCT1 does not interact directly with the phosphate group in the bound phosphopeptide, and its mutation has little effect on sensitivity to DNA damage.
E) Phosphate-binding site mutations in either BRCT1 or BRCT2 domains of Rad4 significantly reduce DNA damage-dependent cell cycle arrest (left), and associated Chk1 phosphorylation and activation by Rad3ATR (middle and right).