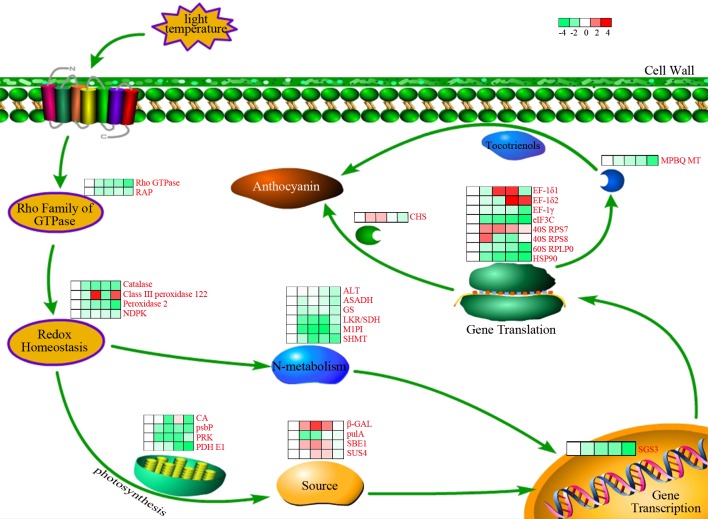
Fig 12. Quantification of black rice grain proteins that are related to ACN biosynthesis.
RAP, Ras-related protein RIC2; NDPK, Nucleoside diphosphate kinase; CA, Carbonic anhydrase; psbP, Chloroplast 23-kDa polypeptide of photosystem II; PRK, Phosphoribulokinase; PDH E1, Pyruvate dehydrogenase E1 component subunit alpha; β-GAL, Beta-galactosidase; pulA, Pullulanase; SBE1, Starch-branching enzyme I; SUS4, Sucrose synthase 4; ALT, Alanine aminotransferase; ASADH, Aspartate-semialdehyde dehydrogenase family protein; GS, Glutamine synthetase; LKR/SDH, Putative lysine-ketoglutarate reductase/saccharopine dehydrogenase bifunctional enzyme; M1PI, Methylthioribose-1-phosphate isomerase; SHMT, Serine hydroxymethyltransferase; SGS3, Protein SUPPRESSOR OF GENE SILENCING 3 homolog; MPBQ MT, 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase 2; EF-1δ1, Elongation factor 1-delta 1; EF-1δ2, Elongation factor 1-delta 2; EF-1γ, Elongation factor 1-gamma; eIF3C, Eukaryotic translation initiation factor 3 subunit C; 40S RPS7, 40S ribosomal protein S7; 40S RPS8, 40S ribosomal protein S8; 60S RPLP0, 60S acidic ribosomal protein P0; HSP90, Hsp90 protein; CHS, Chalcone synthase 1. The protein levels of deregulated enzymes are indicated by colored squares that represent changes in expression (log2 ratio) at various developmental stages in relation to the 3 DAF stage. In a sequence order (left to right), stages are presented from 3, 7, 10, 15, and 20 DAF.