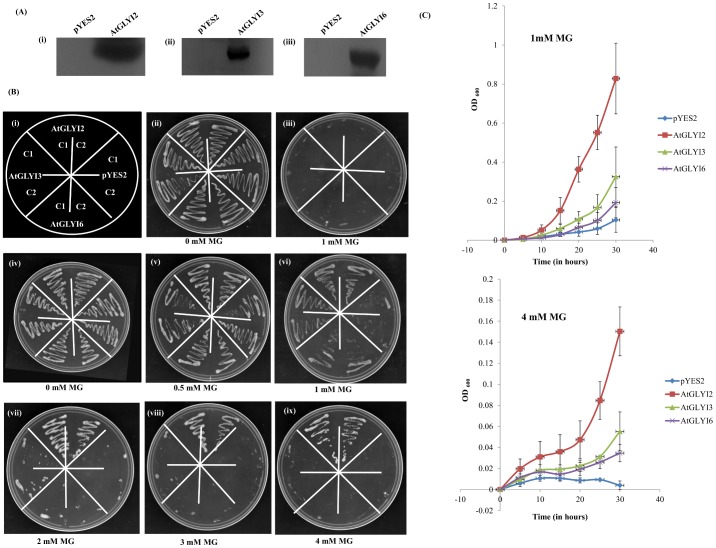
Fig 5. Functional complementation assay using the yeast ΔGLO1 mutant.
The Saccharomyces cerevisiae GLYI mutant cells were transformed with constructs pYES2-AtGLYI2, pYES2-AtGLYI3, pYES2-AtGLYI6 and empty vector pYES2. (A) Western blot depicting expression of AtGLYI2, AtGLYI3 and AtGLYI6 proteins in the total protein extract of yeast cells. Western blotting was done using protein specific antibodies. (B) The transformed cells were grown on solid Ura- SD media with various supplements to check for stress tolerance of yeast cells transformed with AtGLYI genes (i) Pictorial depiction of various strains used (C1 and C2 refers to the two different colonies used). (ii) Medium supplemented with 20% glucose; (iii) Medium supplemented with 20% glucose and 1 mM MG; (iv) Medium supplemented with 20% galactose; (v) Medium supplemented with 20% galactose and 0.5 mM MG; (vi) Medium supplemented with 20% galactose and 1 mM MG; (vii) Medium supplemented with 20% galactose and 2 mM MG; (viii) Medium supplemented with 20% galactose and 3 mM MG; (ix) Medium supplemented with 20% galactose and 4 mM MG. (C) The transformed cells were grown in liquid Ura- SD media supplemented with 20% galactose in presence of 1 mM and 4 mM MG. The growth of cells expressing AtGLYI2 (shown in red color), AtGLYI3 (shown in green color) and AtGLYI6 (shown in purple color) was compared with that of cells expressing empty vector pYES2 (shown in blue color). The growth pattern was monitored spectrophotometrically for 30 h.