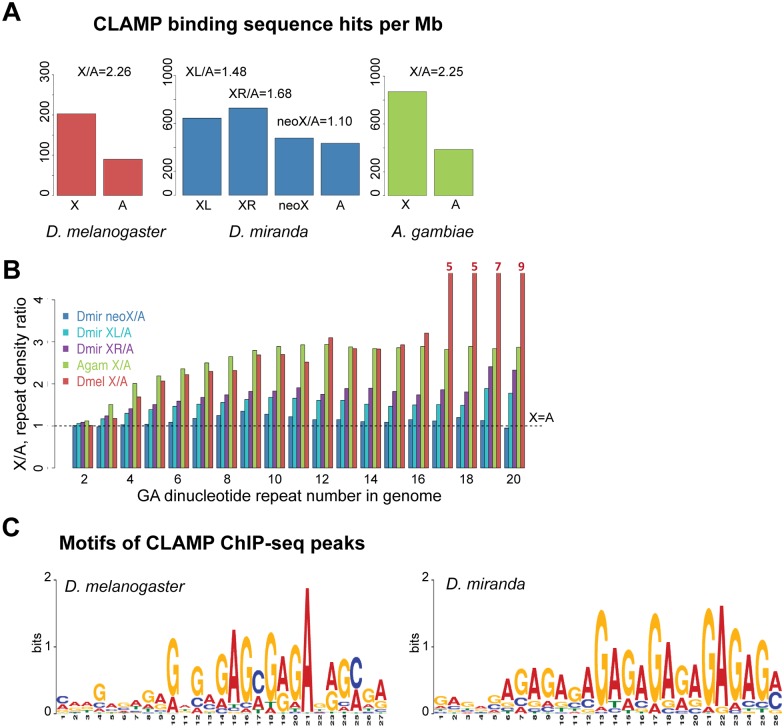
Fig 6. CLAMP binds to similar in vivo X-enriched binding sites in D. miranda and D. melanogaster.
A) The ratio of in vitro CLAMP binding site density (number of CLAMP binding sequence hits per Mb) for X versus autosomes is plotted for D. miranda, D. melanogaster and A. gambiae. Autosomes of D. melanogaster are chromosomes 2L (Muller-B), 2R (Muller-C), 3L (Muller-D), 3R (Muller-E) and 4 (Muller-F); autosomes of D.miranda are chromosomes 2 (Muller-E), 4 (Muller-B) and 5 (Muller-F); and autosomes of A. gambiae are chromosomes 2L (Muller-D), 2R (Muller-E), 3L (Muller-C), and 3R (Muller-B). B) The density ratios of GA-repeats on individual X-chromosome(s) vs. autosomes for different repeat lengths in the D. melanogaster, D. miranda and A. gambiae genomes are plotted. Any value above 1 indicates a higher repeat density on the X-chromosome compared to autosomes. C) CLAMP ChIP-seq motifs are shown for D. melanogaster and D. Miranda larval ChIP-seq data. Motifs are found using MEME-ChIP for the peak regions (peak centers +/-100bp).