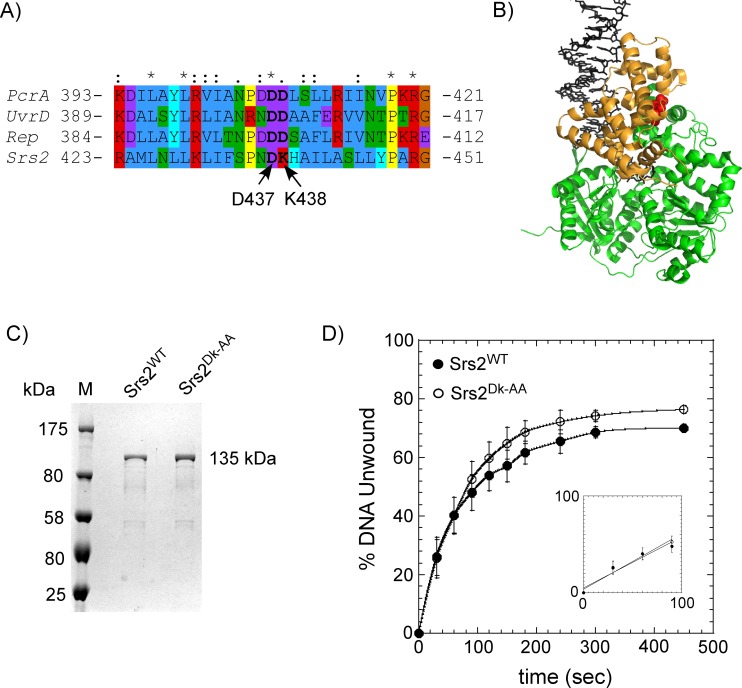
Fig 5. 2B domain mutations in Srs2 have no effect on its DNA unwinding capabilities.
(A) Alignment of the region in the 2B domains from UvrD, Rep, PcrA and Srs2. D437 and K438 in Srs2 align with D403 and D404 in UvrD, which are mutated in the uvrD303 phenotype, a hyperactive helicase mutant of UvrD. Amino acids are colored according to their physicochemical properties. (B) Crystal structure of the UvrD (PDB ID:2IS4; the bacterial Srs2 homolog) is shown with the 2B domain colored gold. The 2B domain is in the ‘closed conformation’ when bound to the unwinding DNA substrate. The DNA is shown as sticks (black) and the D403-D404 residues are shown as red spheres. (C) SDS-PAGE analysis of the purified full length Srs2WT and Srs2DK-AA proteins. (D) Unwinding kinetics of a DNA substrate (25bp dsDNA with a 16 nt 3' ssDNA overhang) by Srs2WT and Srs2DK-AA. No discernable difference in unwinding kinetics is observable between the two proteins. Fitting the linear portion of the data (insert) yield unwinding rates of 0.026 s-1 and 0.028 s-1 for the Srs2WT and Srs2DK-AA proteins, respectively. The mean values and standard errors from three independent experiments are shown.