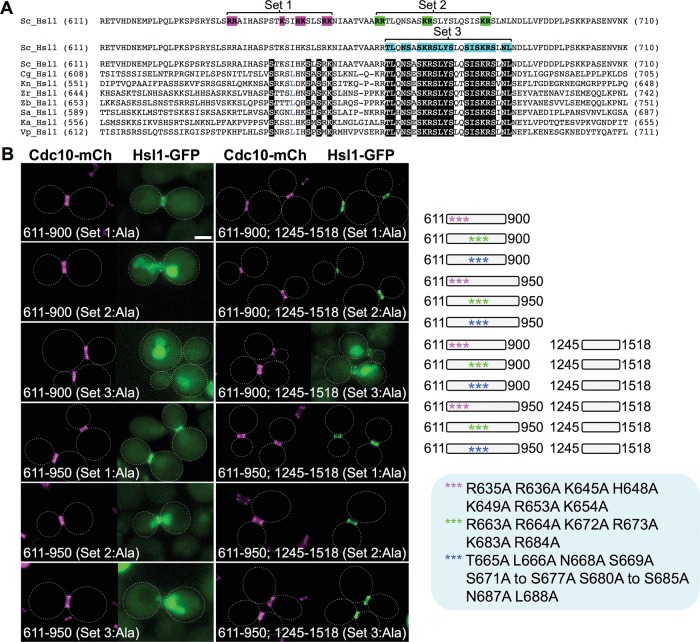
FIGURE 5:
A cryptic bipartite NLS is separable from the conserved septin-associating sequence element in the 611–950 segment of Hsl1. (A) Positions of the residues mutated in the N-terminal portion (residues 611–710) of the 611–950 segment of S. cerevisiae Hsl1 aligned using ClustalW against the corresponding sequences of seven other yeast species (Sc, Saccharomyces cerevisiae; Cg, Candida glabrata; Kn, Kazachstania naganishii; Zr, Zygosaccharomyces rouxii; Zb, Zygosaccharomyces bailii; Nc, Naumovozyma castellii; Ka, Kazachstania africana; and Vp, Vanderwaltozyma polyspora). See also Supplemental Figure S2. White-on-black letters represent invariant residues across all eight fungal species; blue letters indicate strongly conserved residues (found in seven of the eight species). Pink, set 1 allele (R635A R636A K645A H648A K649A R653A K654A); green, set 2 allele (R663A R664A K672A R673A K683A R684A; blue, set 3 allele (T665A L666A N668A S669A S671A K672A R673A S674A L675A Y676A S677A S680A I681A S682A K683A R684A S685A N687A L688A). (B) Right, diagram of the indicated mutated internal fragments, either free or fused to the same C-terminal fragment (and eGFP), that were examined. Bottom, plasmids producing the indicated constructs (pGF-IVL705, pGF-IVL757, pGF-IVL758, pGF-IVL760, pGF-IVL762, pGF-IVL763, pGF-IVL765, pGF-IVL766, and pGF-IVL817 through pGF-IVL820), expressed and visualized as in Figure 1, A and B. Scale bar, 2 μm.