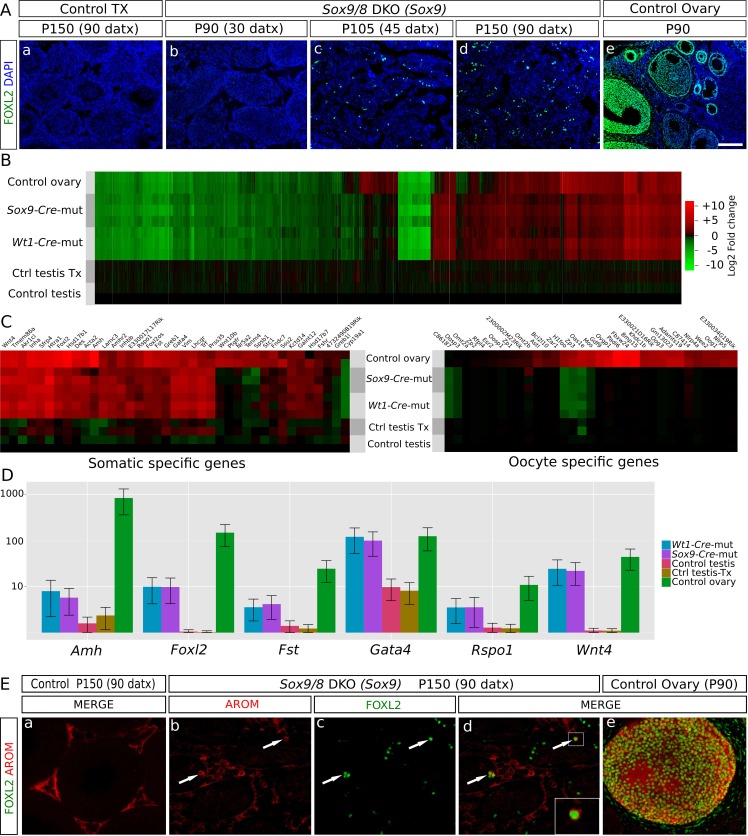
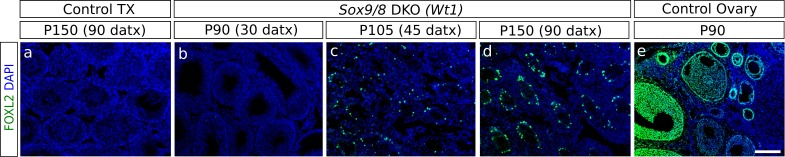
Figure 2. Genetic reprogramming in somatic cells of adult Sox9/8 SC-DKO (Sox9).
(A) Expression of FOXL2 (green fluorescence) in P150 (90 datx) TX-treated control (Sox9f/f;Sox8-/-) (a) and in Sox9/8 SC-DKO (Sox9) mouse testes analyzed at P90 (30 datx) (b), P105 (45 datx) (c), and P150 (90 datx) (d) as well as in a P 90 control ovary (e). (B) Heatmap showing the 12,380 genes found to be differentially expressed at alpha < 0.005 when comparing control (Sox9f/f) and mutant adult gonads. The log2(FPKM+1) of each gene in each condition has been divided by the corresponding value in control testis. Gene expression has not been altered by the TX treatment. Red colors indicate genes upregulated with respect to their expression levels in control testis and green colors indicate downregulated genes. (C) Expression heatmaps of selected ovarian somatic-specific and oocyte-specific genes. (D) Expression bar plots of six relevant ovarian somatic-specific genes upregulated in mutant testes. (E) Aromatase (red) and FOXL2 (green) immunofluorescence staining of TX-treated control (Sox9f/f;Sox8-/-) testis (a), mutant testes (b–d), and control ovary (e). Arrows mark reprogrammed Sertoli cells showing simultaneous expression of Aromatase and FOXL2. Scale bar shown in Ae represents 150 µm in A and 75 µm in E.
DOI: http://dx.doi.org/10.7554/eLife.15635.012