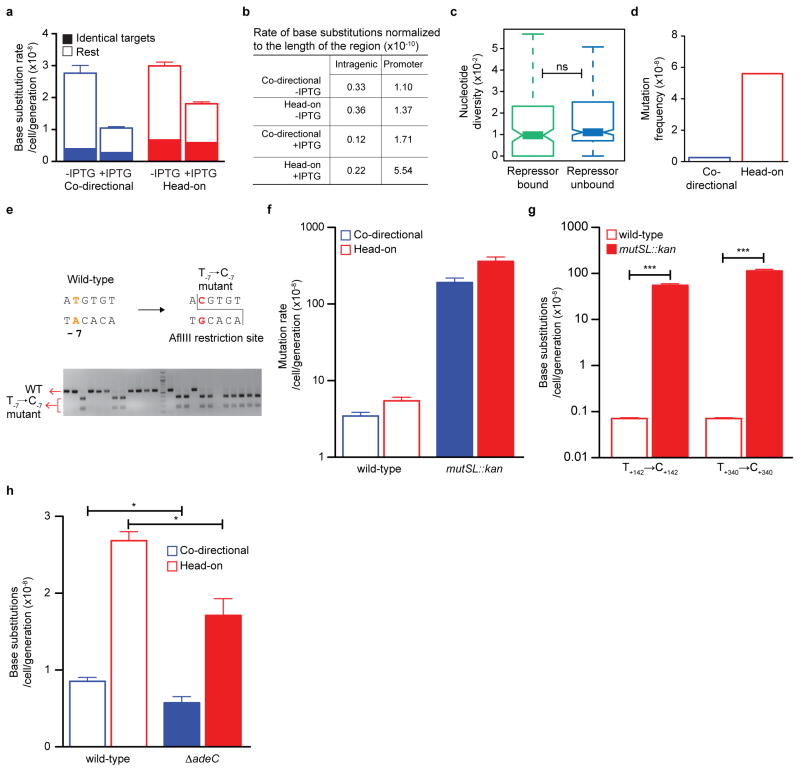
Extended Data Figure 8. Base substitutions and the role of mismatch repair and enzymatic adenine deamination.
a, IPTG-induction does not affect the base substitution rate in coding region of thyP3 when considering identical target sites, indicating that collisions may not be a major source of these mutations. In yeast, it was shown that transcription-associated mutagenesis is proportional to level of transcription10. In B. subtilis, the total rate of base substitutions in the coding region significantly decreases upon IPTG induction, which could be due to an unidentified transcription dependent mutation-correction mechanism, or due to increase of target size of base substitutions in the coding sequence in un-induced (basal) transcription. b, Table showing the rates of base substitutions in coding region and promoter of thyP3 normalized by length of the region. Localized substitution rates are higher in the promoter than coding sequence, thus suggesting that collision has more drastic effect on promoter substitutions. c, Comparative genomic analysis of mutation rates of promoters with and without repressor binding. Nucleotide diversity per site (Theta) was calculated for each promoter across different strains of Bacillus subtilis. The comparison shows no significant difference in nucleotide diversity between repressor-bound promoters and rest of the promoters, indicating that repressor binding may not affect the substitution rate of a promoter. Whole genomes and the repressors analyzed are listed in Extended Data Table 2. (ns-not significant P>0.05; Mann-Whitney U test). d, The mutation frequency of T-7→C-7 mutation is higher in head-on than co-directional orientation in E. coli. The mutation frequency was calculated here from the plasmid-based forward mutation assay data reported by Yoshiyama et al., (2001)26. e, The restriction digestion-based assay to screen for T-7→C-7 mutation. Wild-type promoter sequence does not have an AflIII restriction site, whereas the promoter T-7→C-7 mutation will be digested by AflIII, which is illustrated by a representative agarose gel. f, Mismatch repair mutant (mutSL::kan) shows an expected increase (~60-fold) in total mutation rate of thyP3 in both co-directional and head-on orientation compared to wild-type. The mutation rates of the wild-type strains are presented before in Fig. 1d. g, Mismatch repair mutant shows a drastic ~1000 fold increase in mutation rate of T→C substitution hotspots within the coding sequence of head-on thyP3, indicating that mismatch repair corrects T→C substitution within coding sequence. h, Deletion of adeC gene encoding adenine deaminase modestly reduces the mutation rate of T-7→C-7 substitution in both co-directional and head-on orientation compared to wild-type. For f–h mean±s.e.m of n≥3 experiments is shown. (*P<0.05; ***P<0.001; Student’s t-test).