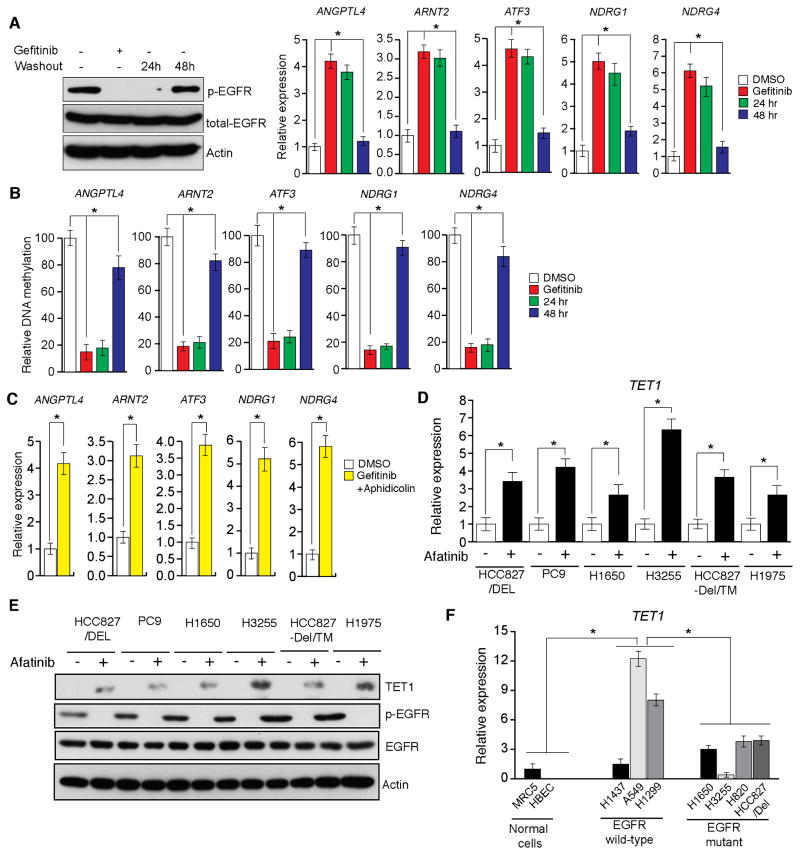
Figure 3. EGFR transcriptionally represses the active DNA demethylase TET1 (See also Figure S2).
(A and B) HCC827/Del cells were treated with gefitinib (0.1 μM) for 48 h and then washed out. DMSO-treated samples, gefitinib-treated samples, and samples harvested 24 and 48 h after washout of gefitinib were collected. (A) Samples were analyzed for the indicated proteins via immunoblot analyses (Left), or for the transcripts of the indicated TSGs via RT-qPCR (Right). (B) DNA methylation of the CpG islands in the promoters for indicated TSGs were analyzed via Me-DIP analyses. The percent of CpG island DNA methylation for HCC827/Del cells under the indicated conditions relative to that of DMSO-treated cells is shown. (C) HCC827/Del cells were treated with aphidicolin (5 μM) and gefitinib (0.1 μM) for 48 h, and the expression levels of the indicated TSGs were analyzed via RT-qPCR. Relative mRNA expression levels for the indicated TSGs in drug-treated cells were compared with those in DMSO-treated cells. (D) The indicated EGFR-mutant lung cancer cell lines were treated with afatinib, and the expression of TET1 mRNA was measured via RT-qPCR. Gene expression was normalized with respect to GAPDH mRNA expression. (E) The indicated EGFR-mutant lung cancer cell lines were treated with afatinib, and the expression levels of TET1 and other indicated proteins were measured via immunoblot analyses. GAPDH expression was used as a loading control. (F) The indicated lung cancer cell lines were analyzed for TET1 expression via RT-qPCR. TET1 expression in lung cancer cell lines relative to that of MRC5 cells is shown. *p < 0.05.