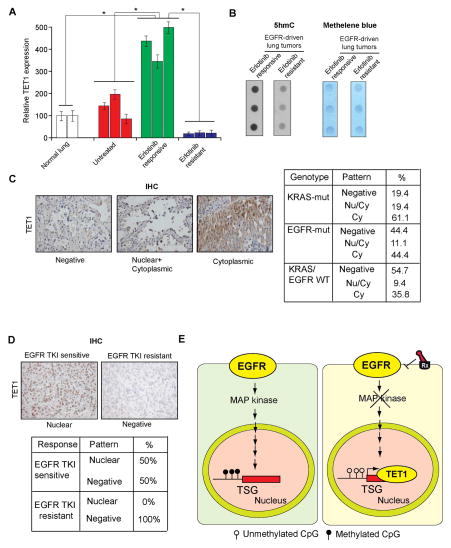
Figure 7. TET1 is downregulated or mislocalized in lung cancer samples and in EGFR TKI resistant samples (See also Figure S6).
(A) Normal lung samples, EGFR-driven untreated lung cancer samples, and EGFR-driven erlotinib-treated lung cancer samples that were either sensitive or resistant to erlotinib treatment were analyzed for TET1 mRNA expression. The relative TET1 expression is shown. (B) Erlotinib-sensitive or erlotinib-resistant lung tumor samples were used to isolate genomic DNA, and the number of 5hmC marks was evaluated by dot blot analyses of 100 ng genomic DNA (Left). The membrane was then stained with methylene blue to confirm equal DNA loading (Right). (C) Analysis of patient-derived lung cancer samples shows the loss of TET1 expression and the cytoplasmic localization of TET1, indicating that TET1 expression is reduced in lung cancer. (Left) Representative images for EGFR-mutant lung cancers and the percent of samples across three genotypes with no staining, cytoplasmic staining, or cytoplasmic and nuclear staining are shown. (Right) Summary of the immunohistochemistry (IHC) results. (D) (Upper panel) Representative images for lung cancer samples stained for TET1. (Lower panel) Summary of the IHC results. (E) The proposed model considers that EGFR represses the active DNA demethylase TET1 to induce epigenetic TSG silencing. TET1 repression is required for EGFR-driven tumor growth and determines the response to EGFR TKIs, because TET1 expression inhibits tumor growth and TET1 knockdown inhibits EGFR TKI-mediated inhibition of cell growth.