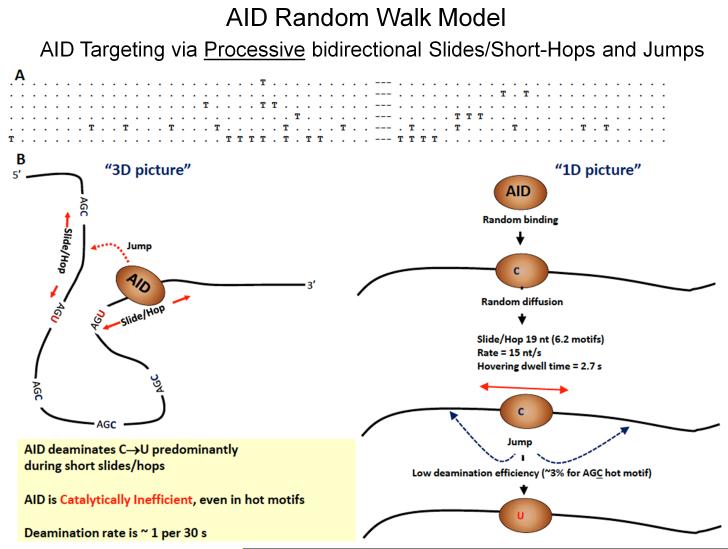
Fig. 9. Random walk model depicting AID scanning and catalyzing haphazard, inefficient deamination of C to U on ssDNA.
A: Representative mutated clones on a cassette containing 32 and 24 consecutive AGC motifs separated by a 9 nt spacer embedded in-frame in a lacZ reporter sequence in an ssDNA gapped region of bacteriophage M13 dsDNA (see text for description). Each T denotes a deaminated WRC motif; each dot denotes a non-deaminated WRC motif; dashes denote 9 nt spacer region. B: AID binds at a random location on ssDNA and scans in random bidirectional short slides/hops. AID only rarely deaminates C → U, at about 3% efficiency even for preferred AGC hot motifs, thus ensuring mutational diversity. The deamination efficiency, slide/hop rates and distances, deamination motif dwell times are predicted from a random-walk mathematical model by an analysis of the types of clonal mutational distributions shown above in A [Mak et al., 2013]. Random deamination clusters can when AID slides repeatedly over the same region of ssDNA. These types of random clustered mutations (termed kataegis) in WRC motifs have been observed as B-cell lymphoma mutational signatures [Qian et al., 2014].