Figure 1. Prevalent nomenclature and timing of macrophages.
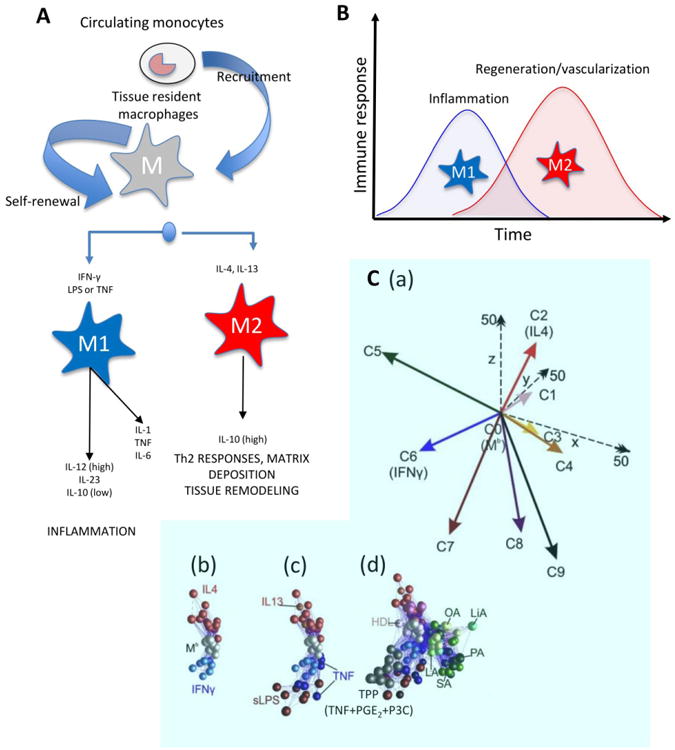
(A) Most adult tissues contain naïve tissue-resident macrophages that originated in the embryonic stage [126]. In addition, circulating monocytes can be recruited to a site of injury where they differentiate into various macrophage phenotypes. According to the most prevalent nomenclature, two macrophage phenotypes have been recognized. The M1 phenotype (classically activated or pro-inflammatory) is activated by IFNγ and LPS. The M2 phenotype (alternatively activated or anti-inflammatory) is activated by IL-4 and IL-13. Modified from Mantovani et al., 2004 [41]. (B) Parallel events occur during bioengineered tissue formation. The biomaterial implantation (with or without cells) will trigger an immune response, which leads to recruitment of various immune cells and macrophage infiltration. At this stage, a dominant M1 macrophage phenotype should be expected, and pro-inflammatory stimuli (required during the first stage of an injury) are upregulated. Next, a timely progression toward an anti-inflammatory chemical scenario could be beneficial for more rapid and smoother implant-tissue integration and healing; thus the M2 phenotype is more desirable in the second stage. (C) A recent report by Xue et al. reported many more than the two widely recognized (M1 and M2) phenotypes: (a) Nine different macrophage phenotypes were identified in human macrophages based on their transcriptome signatures upon activation by different chemical cues. When macrophages were (b) activated with known M1 or M2 chemical cues or (c) with M1 or M2 related chemical signals, they displayed a biochemical behavior consistent with the M1-M2 polarization model. (d) However, activation with other chemical cues (e.g., free fatty acids, high density lipoprotein (HDL), or molecules associated with chronic inflammation) resulted in seven other distinct macrophage phenotypes (a multi-axis spectrum; C1, C3, C4, C5, C7, C8, C9). C2 and C6 are consistent with the expression profile of M2 and M1 phenotypes, respectively. Adapted with permission from Xue et al., 2014 [27].
