FIGURE 2.
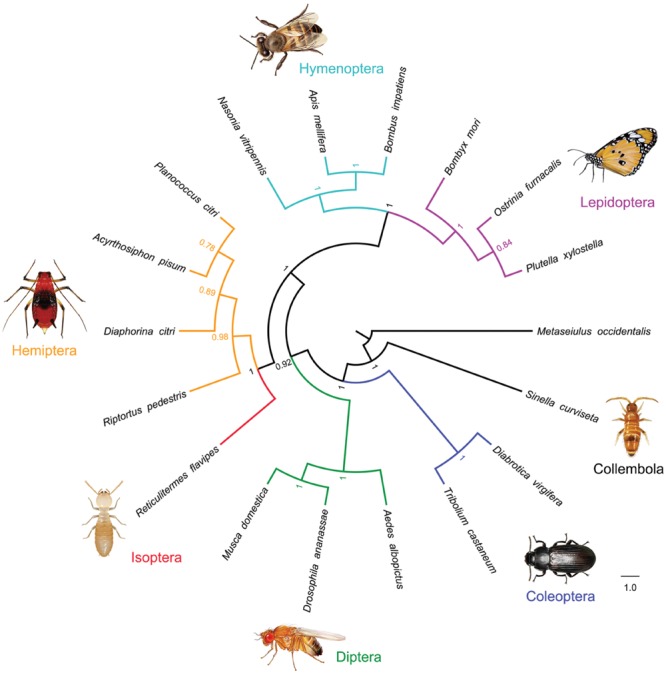
Phylogenetic analysis of v-ATPase A in insects. Amino acid sequences were aligned using the MAFFT algorithm in the TranslatorX online platform. Alignments were then checked and corrected manually in MEGA v6.0. The WAG+G model was the best-fit amino acid substitution model selected by ProtTest 2.4 under AIC criteria. MrBayes v3.2.3 with WAG+G model was used for the phylogenetic analysis.
