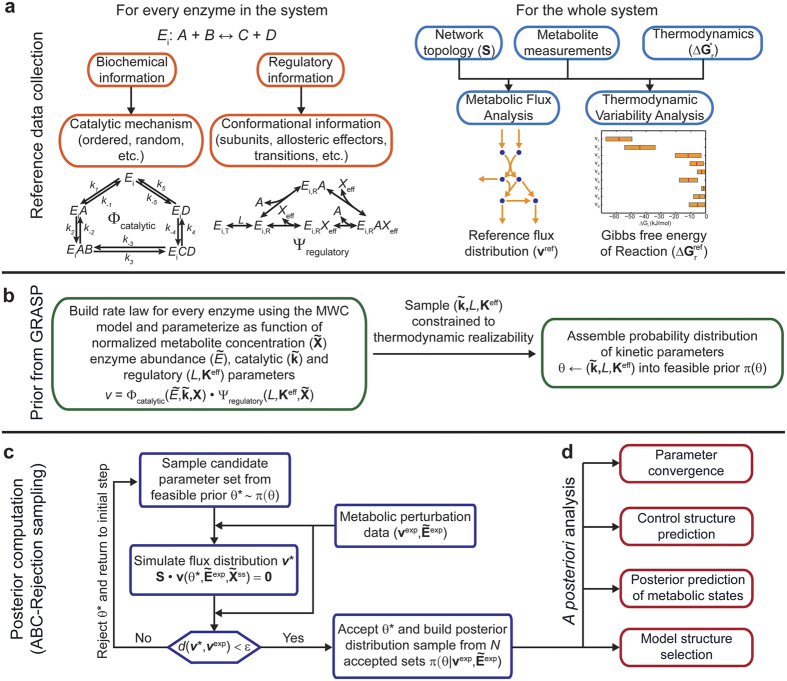
Figure 1. General workflow for building feasible and detailed kinetic models employing a Bayesian approach.
(a) Collection of available biochemical, thermodynamic and structural data for all the components of the system. Reference metabolite concentrations ranges and metabolic fluxes are also needed to define the reference point. (b) Generation of a feasible prior distribution of kinetic parameters (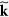 ,L,Keff) using GRASP. This distribution is implicitly described by the auxiliary parameters
,L,Keff) using GRASP. This distribution is implicitly described by the auxiliary parameters  (enzyme intermediate abundances), R (microscopic reversibilities) and relem (branching vector), and spans the full kinetic space allowable by the reaction mechanism, thermodynamics, structural and flux data at the reference state. (c) Computation of the posterior parameter sample is achieved by employing a rejection sampling scheme based on Approximate Bayesian Computation. (d) A posteriori analysis of the posterior distribution enables assessment of parameter convergence, control structure prediction, prediction of metabolic states upon genetic and environmental perturbations, and exploration of possibly missing metabolic interactions.
(enzyme intermediate abundances), R (microscopic reversibilities) and relem (branching vector), and spans the full kinetic space allowable by the reaction mechanism, thermodynamics, structural and flux data at the reference state. (c) Computation of the posterior parameter sample is achieved by employing a rejection sampling scheme based on Approximate Bayesian Computation. (d) A posteriori analysis of the posterior distribution enables assessment of parameter convergence, control structure prediction, prediction of metabolic states upon genetic and environmental perturbations, and exploration of possibly missing metabolic interactions.