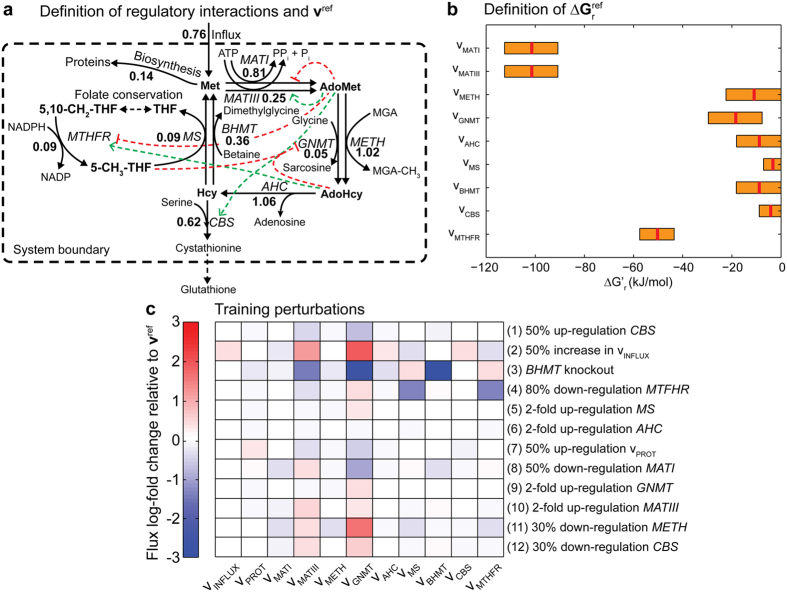
Figure 2. Modelling the mammalian methionine cycle.
(a) Schematic representation of the methionine cycle and its main regulatory interactions. The modelled metabolites are shown in bold, whereas the red and green dashed lines represent known inhibitory (red) and excitatory (green) interactions within this cycle. The enzyme names are displayed in italics. Abbreviations: Met, methionine; AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; Hcy, homocysteine; 5-CH3-THF, 5-methyltetrahydrofolate; 5,10-CH2-THF, 5,10-methylenetetrahydrofolate; THF, tetrahydrofolate; MGA, methyl group acceptor; MGA-CH3 methyl bound to the methyl group acceptor; MATI, methionine adenosyltransferase I; MATIII, methionine adenosyltransferase III; GNMT, glycine-N-methyltransferase; METH, S-adenosylmethionine-dependent methyltransferase; AHC, adenosylhomocysteinase; BHMT, betaine homocysteine S-methyltransferase; MS, methionine synthase; MTHFR, methylenetetrahydrofolate reductase; CBS, cystathionine β-synthase. Bold numbers denote the reference flux distribution of this cycle as calculated by the true model (see Methods). (b) Estimated Gibbs free energy of reactions ranges for each reaction in the cycle. The red lines represent the mean Gibbs free energy of reactions which were used to define the reference thermodynamic point. (c) Heat map of the log-fold change in the flux responses of 12 simulated perturbations relative to the reference flux distribution (a). These experiments represent one-at-a-time genetic and/or environmental perturbations in the reaction fluxes of this cycle. The details of these perturbations are shown in Supplementary Table S1.