Figure 5. The ability of different rsaA constructs to inhibit supercoil diffusion does not correlate with their ability to establish a chromosomal domain boundary.
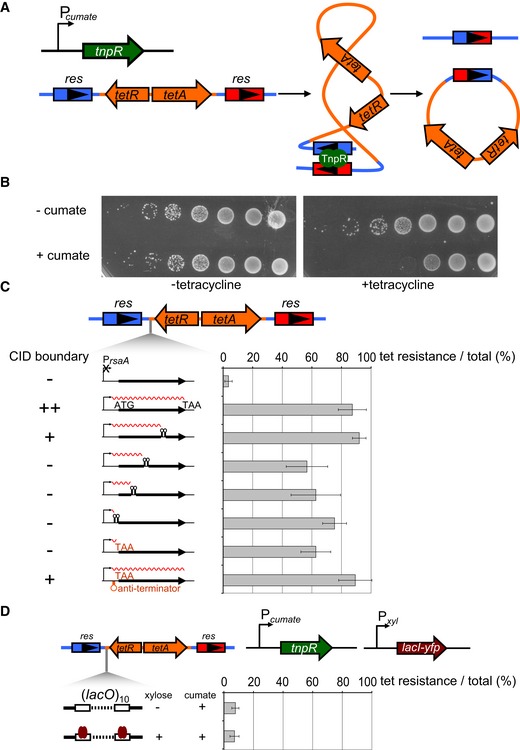
- A γδ TnpR recombinase (green) was induced from a cumate‐inducible promoter. TnpR recombines two res sites (blue and red arrows) that flank a tet resistance cassette (orange arrows) to excise the intervening DNA. Recombination requires supercoiling‐dependent juxtaposition of res sites.
- Induction of TnpR from Pcumate results in a loss of cell viability on tetracycline plates. Each spot is a 10‐fold dilution.
- Percentage of tetracycline‐resistant colonies represent a proxy for the extent of supercoil diffusion inhibition by the various PrsaA‐rsaA constructs indicated, each inserted between one res site and the tet R cassette. Error bars represent SD, n = 3. The ability of each construct to form a CID boundary is indicated on the left (see Figs 2 and 3).
- LacI binding to 10 consecutive lacO sites in between two res sites does not significantly inhibit supercoil diffusion. TnpR and LacI‐YFP (brown arrow) were induced from Pcumate and Pxyl, respectively. Error bars represent SD, n = 3.
