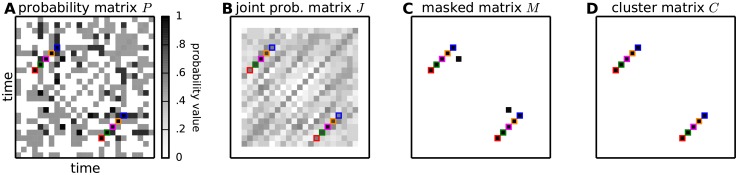
Fig 3. From spike trains to the cluster matrix.
(A) Given parallel spike train data and their intersection matrix I as in Fig 1, for each entry Iij its cumulative probability Pij is calculated analytically under the null hypothesis that the spike trains are independent and marginally Poisson. (B) The l largest neighbors of Iij in a rectangular area extending along the 45 degree direction are isolated by means of a kernel and their joint cumulative probability is assigned to the joint probability matrix J at position Jij. (C) Chosen a significance threshold α1 for the probability of individual entries (i.e. for entries Pij) and a significance threshold α2 for the joint probability of the neighbors of an entry (i.e. for entries Jij), each entry Iij for which Pij > α1 and Jij > α2 is classified as statistically significant. Significant entries of I are retained in the binary masked matrix Mij, which takes value 1 at positions (i, j) where I is statistically significant and 0 elsewhere. (D) 1-valued entries in M falling close-by are clustered together (or discarded as isolated chance events) by means of a DBSCAN algorithm, which thus isolates diagonal structures.