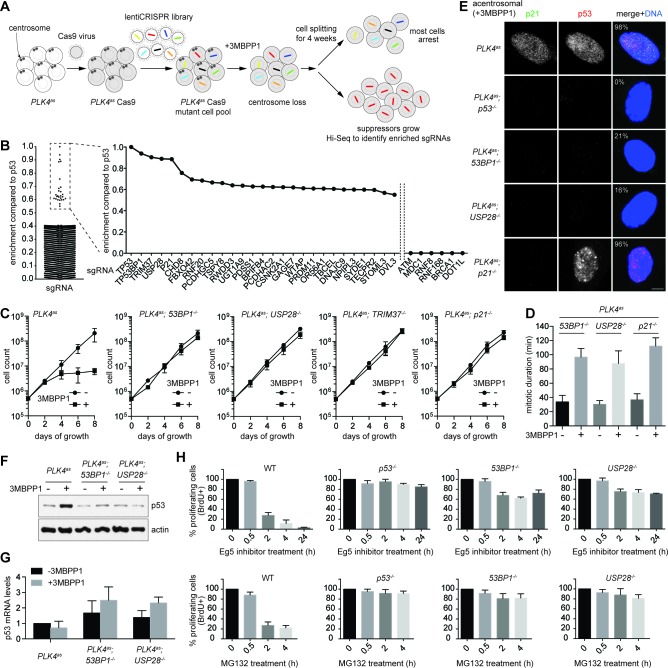
Figure 2. 53BP1 and USP28 are broad acting components acting upstream of p53 in response to mitotic stress.
(A) Schematic representation of the loss-of-function screen for components required for centrosome loss induced G1 arrest using the human genome-scale CRISPR knockout library (GeCKO). (B) Binary logarithm of sgRNA enrichment for genes potentially involving in centrosome loss induced G1 arrest, normalized to the top scoring hit p53. The HiSeq data was collected from eight independent screens. A candidate gene must be hit repeatedly with high scores (HiSeq reads) by two or more of its six sgRNAs in independent screens. Interesting negative hits were also shown, including critical DDR components whose sgRNAs were not enriched but detected only in the baseline reads. (C) Validation of top five scoring hits from the screen other than the p53 control. The results shown here used clonal 53BP1-/-, USP28-/-, and p21-/- CRISPR knockout cell lines derived from PLK4as knock-in cells (PLK4as-KI) obtained from A. Holland (Moyer et al., 2015) (see Materials and methods). The growth curve of the indicated individual CRISPR cell lines in the presence or absence of 3MBPP1 was shown. Data are means ± SD. n>50, N = 3. (D) Acentrosomal 53BP1-/-, USP28-/- and p21-/- cells proliferate in the presence of mitotic delay. Graph showing mitotic duration of the indicated CRISPR cell lines dividing with or without centrosomes measured with live-cell imaging. Data are means ± SD. n>30, N = 3. (E) 53BP1 and USP28 function upstream of p53 to activate G1 arrest. Immunofluorescence images of CRISPR cell lines grown in 3MBPP1 stained with the indicated antibodies. The percentage in the merged panel indicates the proportion of cells with p53 nuclear accumulation. Scale bar, 5 μm. (F) Total p53 levels are not elevated in acentrosomal 53BP1-/-, USP28-/- cells. Immunoblot for p53 protein of the indicated cell lines grown in the presence or absence of 3MBPP1 for seven days. (G) p53 protein elevation during centrosome loss-induced G1 arrest is not due to increased p53 transcription. Quantification of p53 mRNA levels relative to GAPDH in (F) by qRT-PCR. Data are means ± SD. n = 6 from two independent experiments. (H) 53BP1-/-, USP28-/- cells do not arrest in G1 despite experiencing mitotic stresses induced by different drug treatments. BrdU incorporation assay for 24 hr showing proportion of proliferating cells in the indicated CRISRP cell lines following release into mitosis with different duration of Eg5 inhibitor (top panels) and MG132 (bottom panels) treatment and washout. Percentages are normalized to the untreated control. Data are means ± SD. n>250, N = 3.
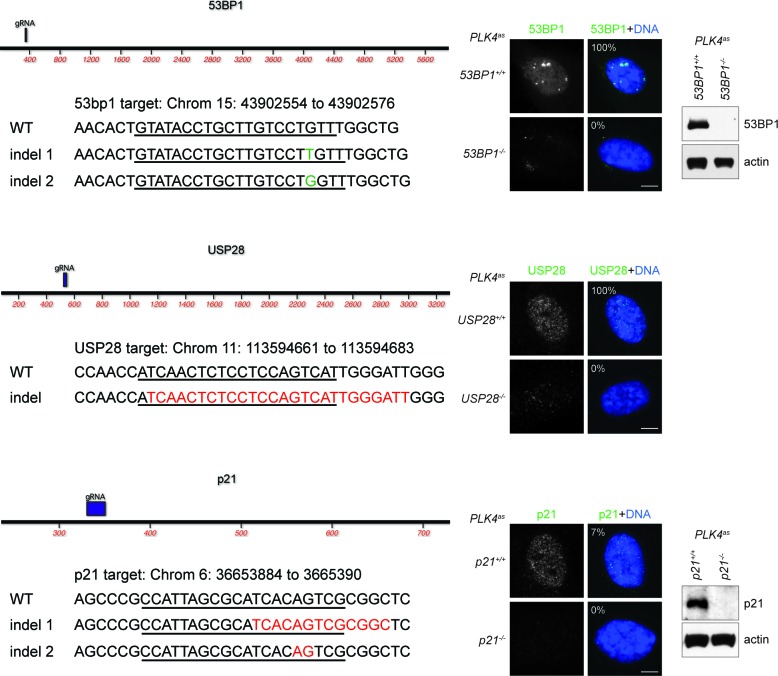
Figure 2—figure supplement 1. Genotyping of CRISPR cell lines.
Figure 2—figure supplement 2. Schematic outlining the timeline of synchronization and drug treatments used.