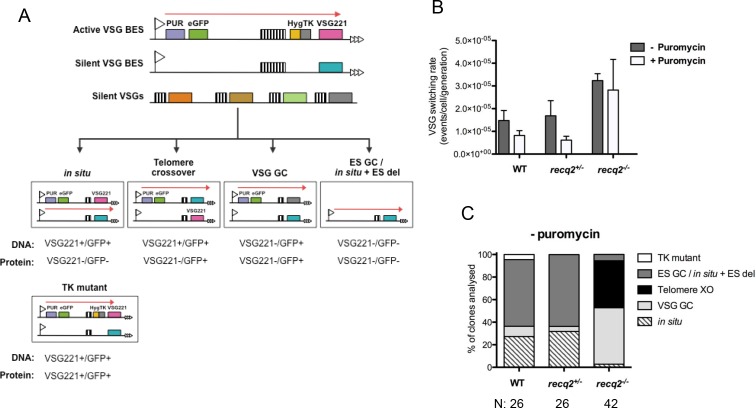
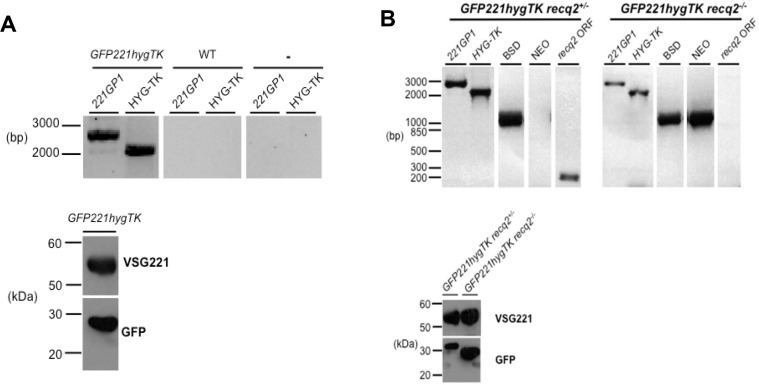
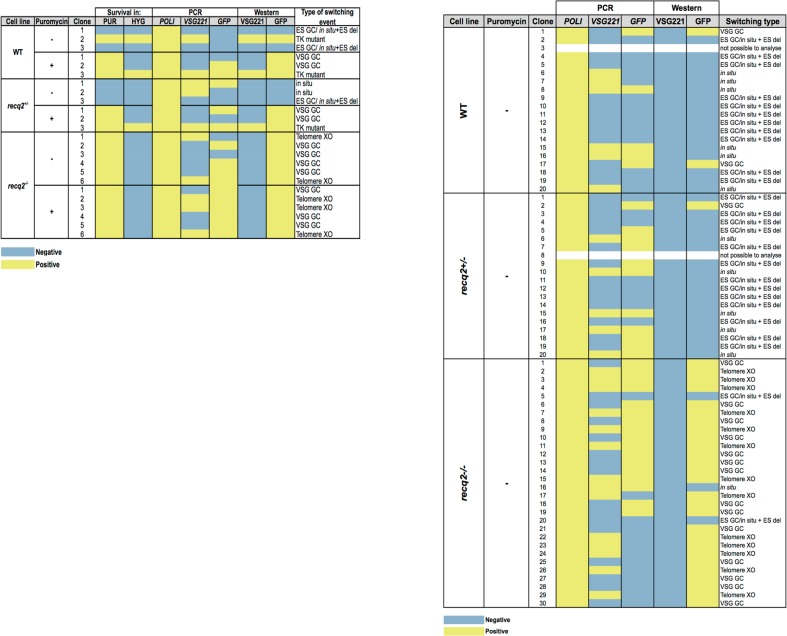
Figure 4. Mutation of TbRECQ2 leads to elevated VSG switching and increased recombination.
(A) Strategy for determining VSG switching mechanisms; adapted from Povelones et al. (2012). The active VSG BES of GFP221hygTK cells is shown, within which the PUR, eGFP, HYG-TK and VSG221 genes are represented as coloured boxes. In addition, one of the ~14 silent BES containing a distinct VSG (turquoise box) is shown, as are multiple silent VSGs elsewhere in the genome (various colours; for convenience these are shown as a single array, but could also be at the telomere of silent mini-chromosomes). 70 bp repeats upstream of the VSGs are denoted by hatched boxes. Different switching strategies allow survival after ganciclovir treatment and can be distinguished by analysis of VSG221 and GFP presence by PCR, and expression of the proteins by western blot (profiles detailed under each mechanism). Switchers that arise by in situ switching, telomere crossover (XO) or VSG gene conversion (VSG GC) can be detected unambiguously, while events that occur by BES gene conversion or in situ switching coupled with BES deletion (ES GC/ in situ+ES del) are indistinguishable. Note, only in situ+ES del reaction is shown, and not ES GC (where all sequence of a silent BES is duplicated and replaces the VSG221 BES); in addition, for VSG GC the silent grey array donor VSG gene is shown as being copied, but the reaction could also use a BES VSG gene. Non-switcher TK mutants can also allow ganciclovir survival. (B) The mean switching rate of GFP221hygTK WT and recq2 mutants (+/- and -/-) was inferred from the mean number of survivors from two experiments, each with three replicates, following treatment with ganciclovir and after culture with (+) or without (-) puromycin; error bars represent standard error of the mean. (C) Profiles of WT and recq2 mutants (+/- and -/-) survivors in the non-puromycin experiments, represented as a percentage of total surviving clones analysed from the two datasets; number of clones (N) analysed is indicated.