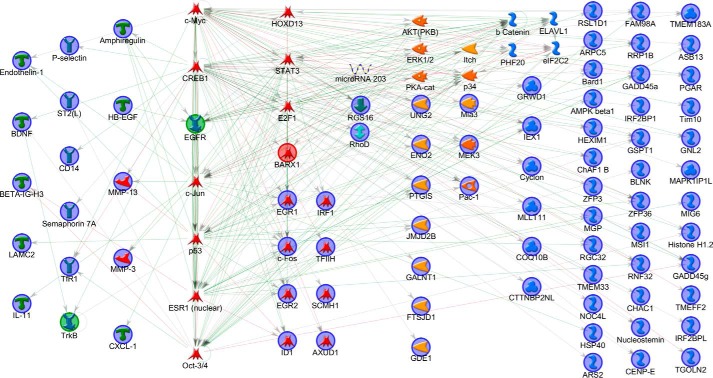
FIGURE 7.
Network of transcriptional regulation and ligand receptor signaling. This network uses a greater number of genes, including all those identified as significantly changed by combining significantly differentially expressed genes with those significantly up-regulated at both time points. The network was generated in MetaCore using the following options: AN(TF)-building algorithm with “add ligands and TF targets” selected. The algorithm generates a list of possible networks, with scores based on the number of seed nodes to non-seed nodes and the presence of canonical pathway threads. The network with the highest score for these two factors was selected and manually organized, first showing the four validated genes and then the most highly connected objects regardless of functional type to their right. The remaining objects were sorted by function so that TFs, kinases, phosphatases, generic proteins, and binding proteins were from left to right. Ligands and receptors were placed to the far left. Seed nodes are circled in navy blue, the predicted receptor trigger is highlighted in green, the predicted controlling TF is highlighted in red, lines represent interactions either transcriptional regulation or protein-protein associations; red = inhibition and green = activation.