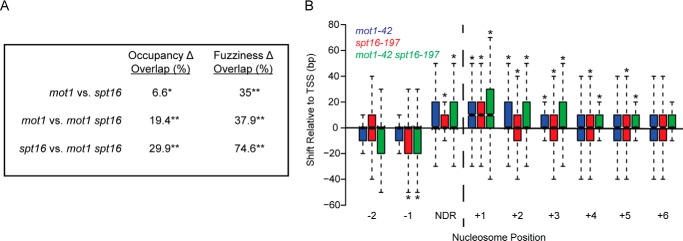
FIGURE 10.
Overlapping sets of nucleosomes are affected in mutant strains. A, the table shows the overlap in sets of nucleosomes affected in mot1-42, spt16-197, and double mutant cells. False discovery rate-corrected p values are <0.007 (*) and <2.6 × 10−196 (**). B, differentially fuzzy nucleosomes in mutant strains were analyzed for position with respect to the TSS. Analysis of the results in Fig. 9 established that the +1 nucleosome was located from approximately −50 to +100 bp with respect to the TSS. Nucleosomes downstream of the +1 nucleosome were assigned to non-overlapping 150-bp bins based on the +1 nucleosome position. The distance from the nucleosome dyad to the TSS was then determined for each nucleosome and in each strain. The plot shows the distribution of median differences in position with respect to the TSS for each nucleosome in the indicated mutant compared with WT. The +1, +2, and +3 nucleosomes in mot1-42 (blue), spt16-197 (red), and mot1-42 spt16-197 (green) cells had significant shifts away from the TSS. The −1, +4, and +5 nucleosomes all shifted away from the TSS in spt16-197 and double mutant cells. Asterisks indicate p < 0.05 as determined by a Kolmogorov-Smirnov test. NDR, nucleosome-depleted region.