FIGURE 6.
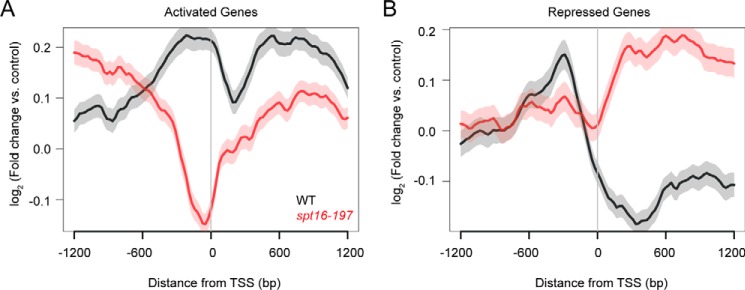
Effect of Spt16 mutation on gene average localization of Mot1. A, Mot1 ChIP-Seq signal was determined as log2 -fold change from two replicates relative to an input control. Genes were computationally aligned by TSS (0 bp) to determine average localization in WT (black) and spt16-197 (red) strains. In spt16-197 cells, Mot1 ChIP signal decreased most dramatically in the promoter region (∼−300 to 0 bp), consistent with decreased expression of the Mot1 and Spt16 activated gene class. There were also changes in Mot1 signal upstream and downstream of the promoter region in the absence of Spt16, suggesting that Spt16 establishes a chromatin environment important for Mot1 binding. B, plot similar to that in A showing the Mot1 localization at Mot1 and Spt16 co-repressed genes. Mot1 ChIP signal increased in the ORF in the absence of Spt16, potentially due to exposed adventitious binding sites for TBP (and hence Mot1). In WT cells and for both classes of genes, Mot1 was notably enriched in the promoter region as expected. In A and B, the S.E. is shown by the transparent shading around the lines.