FIGURE 7.
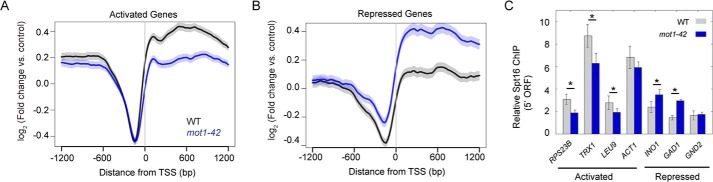
Gene average chromatin localization of Spt16 and dependence on Mot1. A, average Spt16 ChIP-Seq signal in WT (black) and mot1-42 (blue) strains. At Mot1 and Spt16 co-activated genes, Spt16 was depleted from regions upstream of the TSS (0 bp) but was enriched in flanking transcribed regions. Spt16 levels decreased in the ORFs in mot1-42 cells, consistent with the decreased expression of these genes. B, in mot1-42 cells, Spt16 signal increased in the transcribed regions of co-repressed genes, consistent with their increased expression. In A and B, the S.E. is shown by the transparent shading around the lines. C, Spt16 ChIP at selected co-regulated genes. In mot1-42 cells, Spt16 ChIP signal decreased at co-activated genes and increased at co-repressed genes, consistent with changes observed in A and B and RNA levels (Fig. 3, C and D). Error bars are one standard deviation from the mean from at least three biological replicates, and asterisks denote p < 0.05.