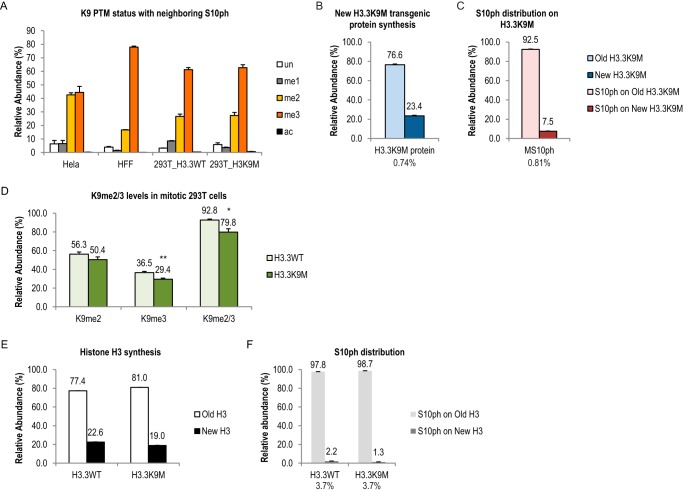
FIGURE 4.
Neighboring H3K9 was not required for the asymmetric distribution of Ser-10 phosphorylation. A, H3S10ph frequently coexisted with K9me2/3 in mitotic human cells. Bar graphs show relative abundance of H3K9 PTM statuses with a neighboring Ser-10 phosphorylation. HeLa cell data were collected from three pulse-SILAC experiments with four time points. The error bars show standard error of biological replicates. HFF cell data were collected from two pulse-SILAC experiments. 293T cells were collected from one pulse-SILAC experiment. The error bars show standard error of multiple MS runs. B and C, H3K9 residue was not required for asymmetric distribution of its neighboring Ser-10 phosphorylation on a transgenic H3.3K9M protein in 293T cells. Bar graphs show H3.3K9M protein synthesis (B) and distribution of Ser-10 phosphorylation on the mutant protein (C). The number below B shows level of H3.3K9M protein is 0.74% of total H3. The number below C indicates 0.81% of the mutant protein had Ser-10 phosphorylation. The numbers above each bar show average of three MS runs. Error bars show standard error. D–F, H3S10ph level and distribution remained the same in 293T cells expressing H3.3K9M transgenic protein. Bar graphs showing relative abundance of the following items in 293T cells expressing either HA- and FLAG-tagged H3.3 wild type (WT) protein or H3.3K9M mutant protein. D, K9me2/3; E, new and old WT H3 protein, including endogenous protein; F, Ser-10 phosphorylation on old and new H3. The numbers above each bar show the average. Error bars show standard error. The numbers below (F) show the percentage of Ser-10 phosphorylation from the wild type H3 protein. *, p < 0.05; **, p < 0.01.