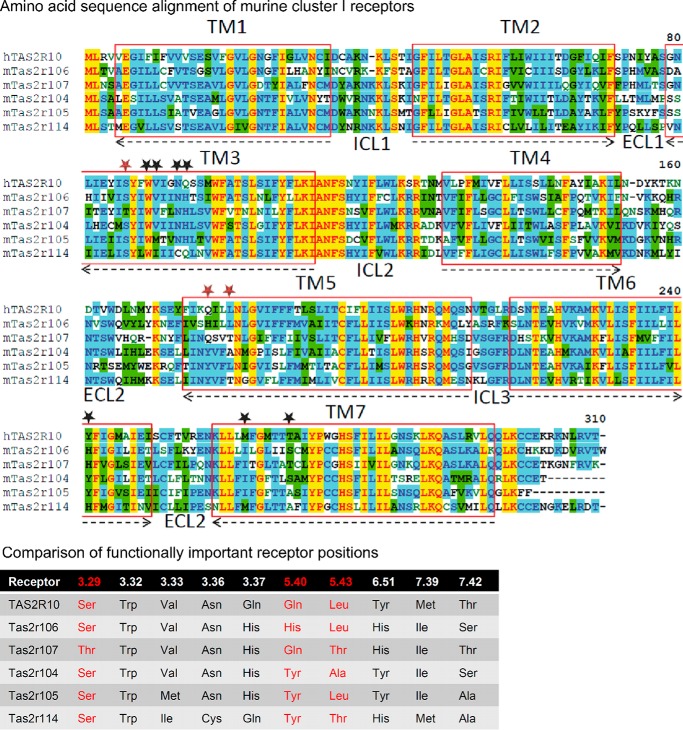
FIGURE 10.
Sequence comparison of muroid cluster I. Upper panel, amino acid sequence alignment of muroid cluster I receptors. The alignment was created using the AlignX program of the Vector NTI software (Life Technologies). Residues are labeled according to the degree of conservation: yellow, identical; green, conserved; blue, identical in at least half of the sequences. The transmembrane domains (TM) are labeled by red boxes and numbered. The orientation in the lipid bilayer is shown by arrows (arrowheads point toward the extracellular site). Intracellular (ICL) and extracellular loops (ECL) are indicated and numbered. Amino acid positions demonstrated to reside in the ligand binding pocket of human TAS2R10 are indicated by stars. Red stars highlight amino acid positions with pronounced agonist selectivity, and black stars contribute to ligand binding and binding pocket constitution (30). Lower panel, comparison of functionally important receptor positions. The six receptors belonging to muroid cluster I were compared for their conservation at positions experimentally proven to contribute to agonist binding in human TAS2R10, a member of this cluster. Receptor residues are indicated by their position according to the Ballesteros-Weinstein numbering system (124). Red-labeled residues reside in positions exhibiting pronounced agonist selectivity in TAS2R10; other positions were demonstrated to constitute the receptor binding site (30).