Figure 5. Autoubiquitination of Hrd1 is required for retrotranslocation.
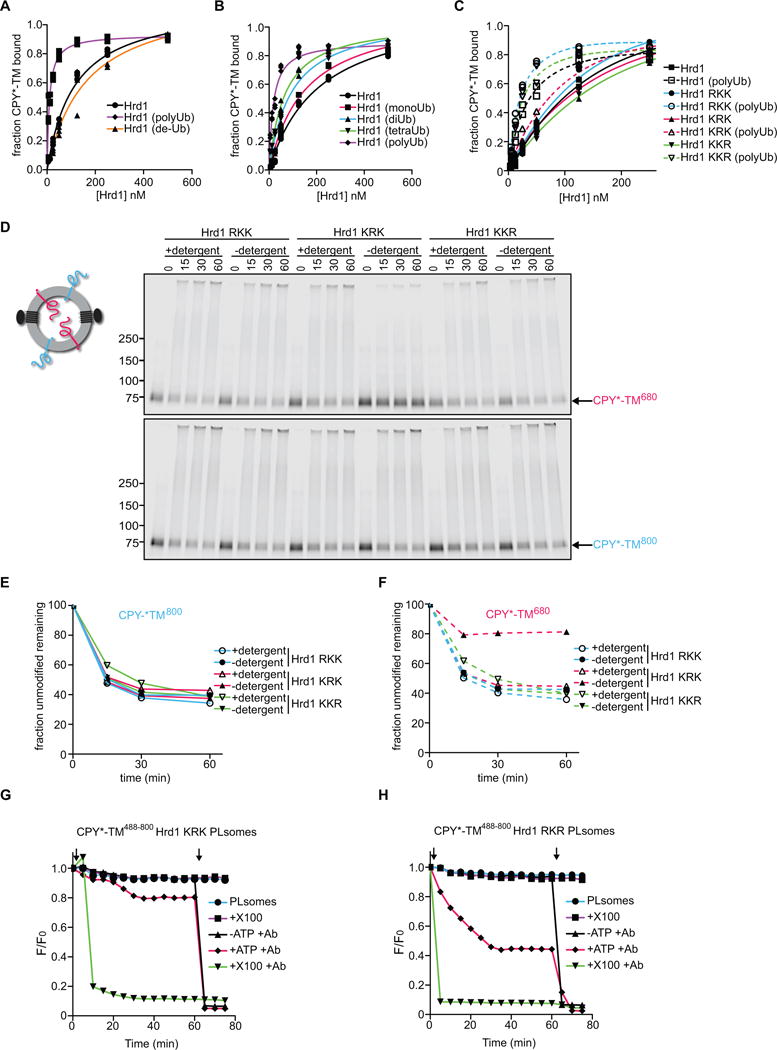
(A) DyLight800-labeled CPY*-TM was incubated with different concentrations of unmodified Hrd1, pre-polyubiquitinated Hrd1 (poly-Ub), or polyubiquitinated Hrd1 treated with the de-ubiquitinating enzyme Usp2 (de-Ub). After incubation with streptavidin beads, the amount of bound material was analyzed by SDS-PAGE and fluorescence scanning. Note that modified Hrd1 has a higher affinity for substrate.
(B) As in (A), but Hrd1 was also pre-modified with mono-, di- or tetra-ubiquitin.
(C) As in (A), but Hrd1 variants were used, which contained lysine-to-arginine substitutions in either the transmembrane domain (RKK), the RING-finger domain (KRK), or the C-terminal tail (KKR). In each case, Hrd1 was tested with and without polyubiquitination. Note that the ubiquitination-mediated increase of substrate affinity is blunted for the KRK mutant.
(D) Proteoliposomes containing substrate in both orientations and one of the Hrd1 mutants described in (C) were incubated with ubiquitination machinery for different time periods in the presence or absence of detergent. The samples were analyzed by SDS-PAGE and fluorescence scanning. Note that the CPY* domain of the red molecules is not efficiently retrotranslocated by the KRK mutant.
(E) Quantification of the fluorescence at 800 nm in (D).
(F) Quantification of the fluorescence at 680 nm in (D).
(G) Retrotranslocation of CPY*-TM was tested with the fluorescence quenching assay as in Figure 3H, but with Hrd1 carrying lysine to arginine substitutions in the RING domain (KRK mutant).
(H) As in (G), but with Hrd1 containing lysines only in the RING domain (Hrd1 RKR).
See also Figure S5.
