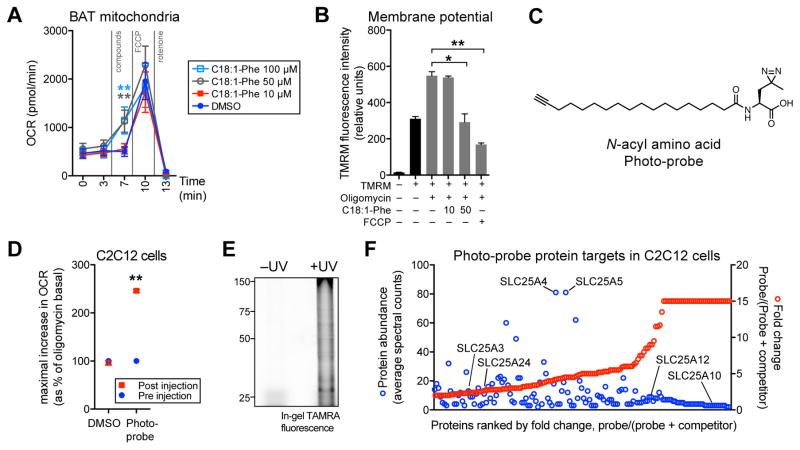
Figure 6. Effects of N-acyl amino acids in mitochondria and identification of N-acyl amino acid-interacting proteins.
(A) Oxygen consumption rates (OCRs) of freshly isolated BAT mitochondria treated with the indicated compounds for the indicated times. Respiration was measured with 10 mM pyruvate and 5 mM malate as substrates, and FCCP and rotenone were used at 2 μM and 3 μM, respectively. n=4–5/group, ** p<0.01.
(B) Tetramethyl rhodamine methyl ester (TMRM) fluorescence in C2C12 cells following 20 min treatment with oligomycin alone (1 μM), or in combination with C18:1-Phe (10 or 50 μM) or FCCP (0.4 μM). n=3/group, mean ± SEM, * p<0.05, ** p<0.01.
(C) Chemical structure of the N-acyl amino acid photocrosslinkable probe (“photo-probe”).
(D) OCR of C2C12 cells treated with DMSO or the photo-probe (50 μM). For (D), data is shown as the maximal increase in OCR as a percentage of the oligomycin basal OCR, which is normalized to 100%. n=3–4/group, mean ± SEM, ** p<0.01
(E) TAMRA in-gel fluorescence of C2C12 cells treated with the photo-probe (50 μM, 20 min), followed by UV irradiation (on ice, 10 min), cell lysis, and click chemistry with TAMRA-N3. For (E), control cells that were not UV irradiated were kept under ambient light (on ice, 10 min).
(F) Proteins in C2C12 cells that showed C20:4-Phe competeable photoprobe labeling. For (F), C2C12 cells were incubated with 20 μM photo-probe (“probe only”), or 20 μM photo-probe with 100 μM C20:4-Phe competitor (“probe + competitor”). Cells were then UV irradiated, lysed, subjected to click chemistry with biotin-N3, and analyzed by MS (see Methods). Proteins satisfying the following filtering criteria are shown: >50% reduction in peptide counts with competitor present versus without competitor, and detection of at least one peptide in all three probe only samples. Comparisons in which no peptides were detected in “probe + competitor” samples were assigned a fold-change of 15.
See also Table S5.