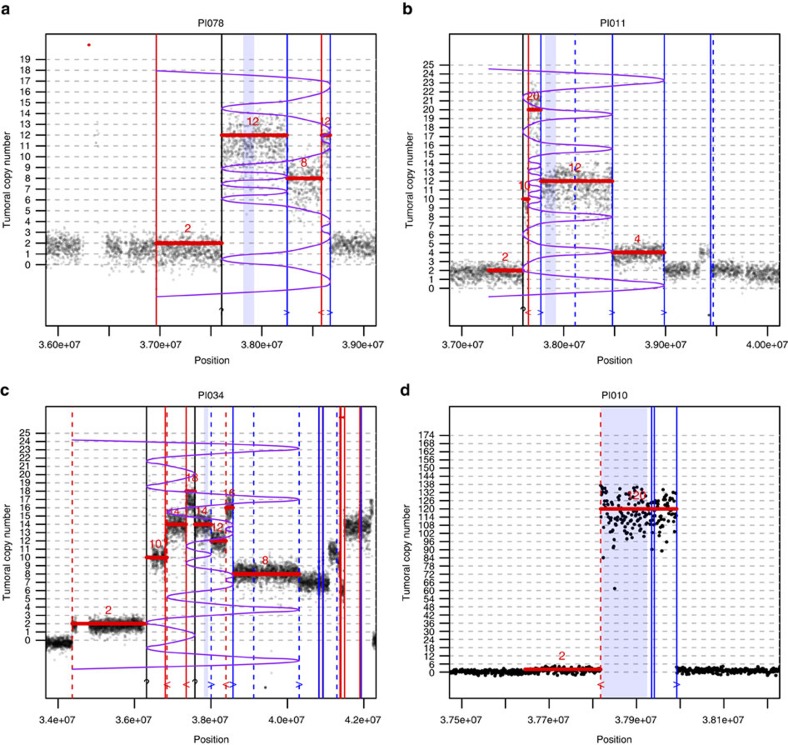
Figure 2. Examples of CNV patterns in the ERBB2 amplicon.
X axis: position on chromosome 17, the minimal region defining the ERBB2 amplicon is indicated by the blue shaded box. Y axis: tumoral integer copy number, computed in 1 kb binned read counts (Methods). Horizontal red segments indicate the closest sequence consistent with a breakage−fusion−bridge (BFB) fold and the purple curved line represents the corresponding folding pattern. Vertical lines indicate the location of detected breakpoints from discordant and clipped read pairs (Methods); plain lines correspond to intra-chromosomal events and dashed lines to inter-chromosomal events; line colour (and glyph on the bottom) correspond to the clipped reads orientation, red (<): reads clipped to the left, blue (>) reads clipped to the right. If the predicted BFB folding pattern is correct, clipped reads orientation should correspond to a left (resp. right) fold in the purple line. Black lines correspond to missed breakpoints, which (approximate) position could be inferred from a copy-number change. (a) A BFB consistent sequence composed of intra-chromosomal events (plain lines) only. (b) Same as previous with additional inter-chromosomal events (dashed lines) not involved in BFB. (c) A BFB consistent sequence composed of both intra- and inter-chromosomal events. The presence of inter-chromosomal events suggests that the amplification process may involve other chromosome(s). (d) Example of a focal amplification, unlikely to occur by a BFB mechanism, suggesting that other mechanisms (double minutes) may be involved.