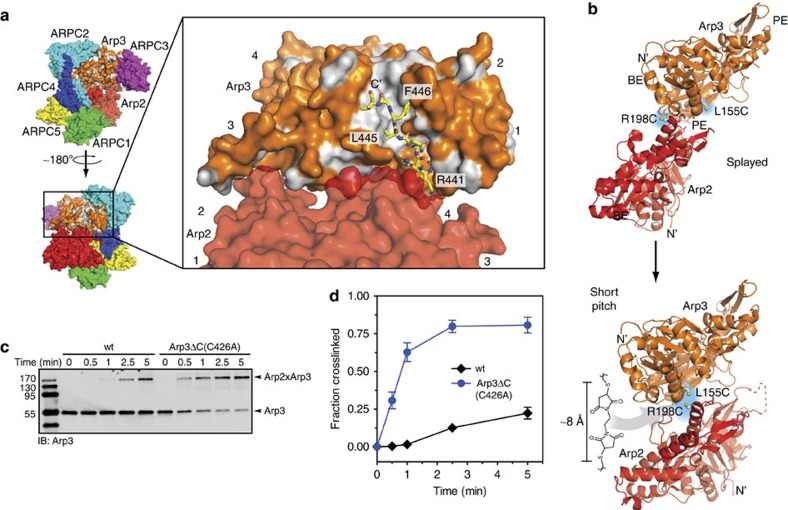
Figure 4. The Arp3 C terminus is required to prevent formation of the short-pitch conformation.
(a) Surface representation of BtArp2/3 complex (4JD2) showing the barbed end of Arp3, the barbed end groove and Arp3 C-terminal tail (yellow stick representation). Residues in Arp3 are coloured according to hydrophobicity, with polar residues orange and non-polar residues grey. Residues are labelled based on the S. cerevisiae Arp2/3 complex sequences. (b) Ribbon diagrams of Arp2 and Arp3 showing the position of engineered cysteines (Arp3(L155C), Arp2(R198C)) in the splayed and short-pitch conformations and the structure of the chemical crosslinker (BMOE) used in the short-pitch crosslinking assay. Structure 4JD2 was used to make both panels. In the right panel, the Oda et al.68 actin filament structure was used to move Arp2 into the short-pitch conformation. The solvent accessible crosslinking distance between engineered cysteines is 32.5 Å in the splayed conformation, and ranges from ∼8 to 11.3 Å in different models of the short-pitch conformation (Supplementary Fig. 3)43. BE: barbed end; PE: pointed end. (c) Anti-Arp3 western blot of 1-min crosslinking reactions containing 1 μM wild type (WT) or Arp3ΔC ScArp2/3 complexes in 200 μM ATP, 1 mM MgCl2, 50 mM KCl, 10 mM Imidazole pH 7.0, and 1 mM EGTA. Both complexes harbour the dual-engineered cysteine residues. The Arp3ΔC complex also contains the C426A mutation to eliminate the non-short-pitch crosslinking product (Fig. 7). (d) Quantification of reaction described in c. Error bars show s.e. for three reactions.