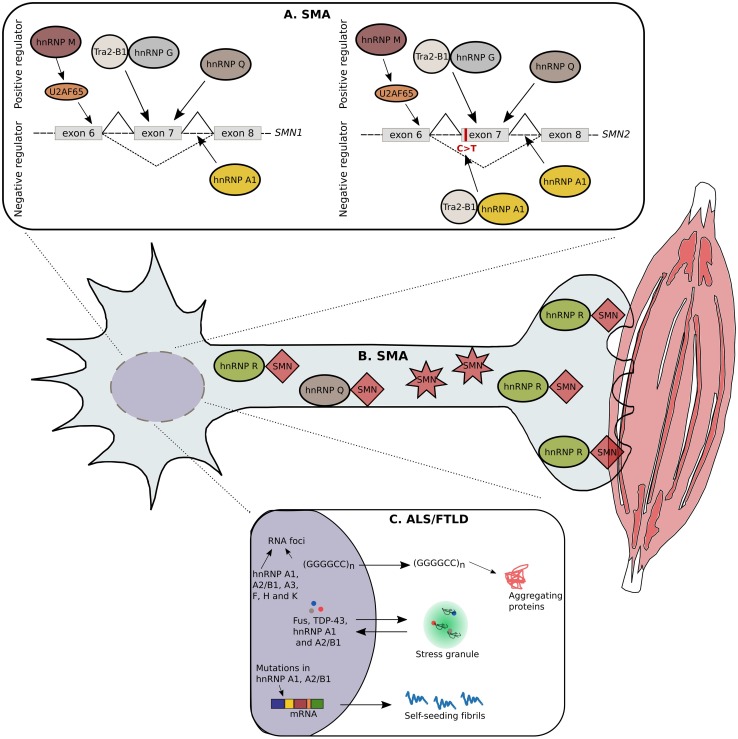
Fig. 5.
hnRNPs involved in SMA and ALS/FTLD. Due to extensive research in the last years, numerous hnRNPs are linked with SMA and ALS/FTLD. A single nucleotide substitution in the 5′-end of exon 7 in the SMN2 gene leads to exon skipping and consequently to a non-functional SMN protein. The hnRNP G can form a complex with the positive modifier, Tra2-β1, resulting in the retention of exon 7. In addition, hnRNP Q can bind to the single nucleotide substitution, thereby avoiding exon 7 skipping. The hnRNP M targets a splicing enhancer on exon 7 through the recruitment of U2AF65, leading to the production of a full-length transcript. In contrast, hnRNP A1 is found to be a negative regulator by binding to the splicing silencer, created by the single nucleotide substitution in SMN2. In addition, hnRNP A1 can bind to regions located in the introns of SMN1/2 (a). Besides being involved in alternative splicing, the hnRNP Q forms protein interactions with wild-type SMN, but is unable to bind to the truncated form. Similarly, the hnRNP R interacts with SMN in the cytosol of motor neurons; more specifically this interaction is present in the presynapses of neuromuscular junctions (b). C9orf72 repeat expansions causing ALS/FTLD leads to the formation of RNA foci containing hnRNP A1, A2/B1, A3, F, H and K suggesting RNA toxicity via the sequestration of RBPs as a possible pathomechanism. Furthermore, hnRNP A1 and A2/B1 are sequestered in cytosolic stress granules together with other RBPs causative for ALS/FTLD, such as FUS and TDP-43. Mutations found in the prion-like domain of hnRNP A1 and A2/B1 are causative for ALS/FTLD. In normal conditions, these hnRNPs have an intrinsic tendency to self-aggregate, because of their prion-like domains, but this tendency is abnormally increased because of the disease-causing mutations (c)