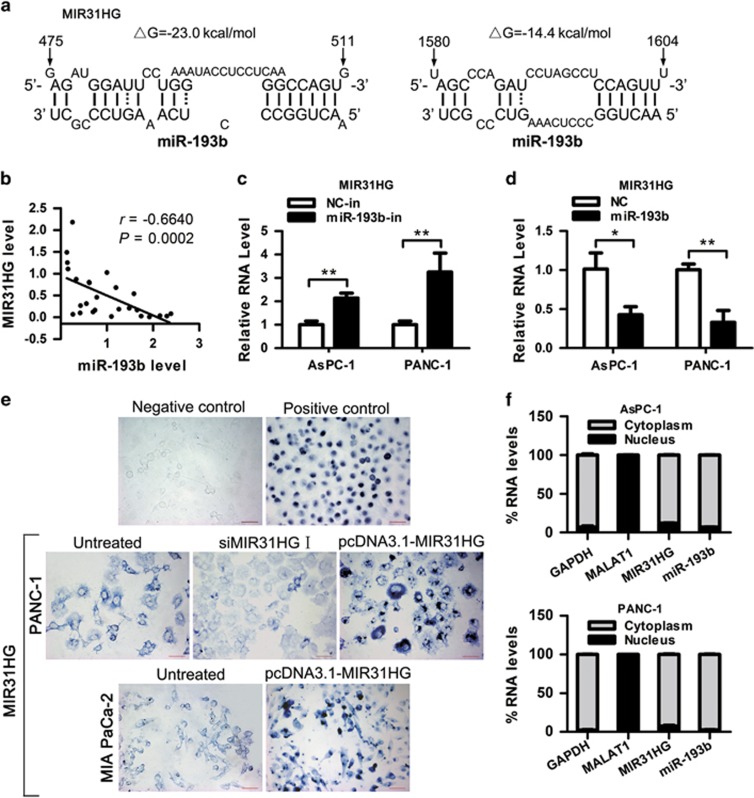
Figure 5.
Negative regulation of MIR31HG by miR-193b. (a) Predicted binding sites for miR-193b in MIR31HG sequences. Numbers show the nucleotides relative to the transcriptional start site of MIR31HG. (b) The expression of MIR31HG was negatively correlated with that of miR-193b (r=−0.6640, P=0.0002) in clinical specimens. (c, d) AsPC-1 and PANC-1 cells were transfected with miR-193b inhibitor (c) or mimic (d), and 48 h later total RNA was isolated and the expression of MIR31HG was analyzed by RT–qPCR. *P<0.05, **P<0.01. (e) RNA ISH assay of MIR31HG in PDAC cells. The blue signal is from the digoxigenin-labeled RNA probe. Negative control (Sense MIR31HG probe) shows no staining and positive control (MALAT1 probe) shows nuclear blue staining in PANC-1 cells. Scale bars, 50 μm. (f) RT–qPCR detection of the percentage of MIR31HG, miR-193b, GAPDH and MALAT1 in the cytoplasmic and nuclear fractions of AsPC-1 and PANC-1 cells. GAPDH and MALAT1 serve as a cytoplasmic and nuclear localization marker, respectively. Abbreviation: ISH, In situ hybridization.