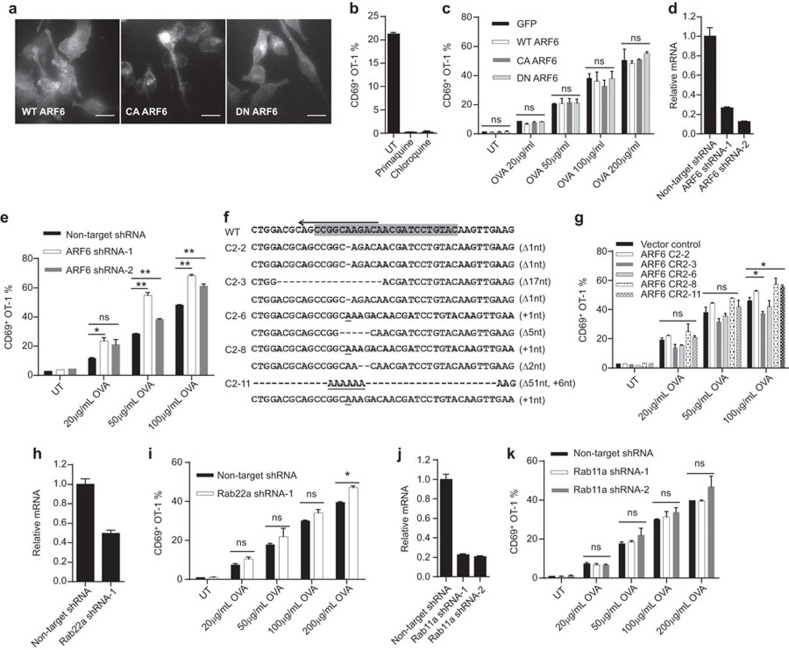
Figure 1.
Disruption of key GTPase regulators of endocytic recycling compartment does not interfere with soluble antigen crosspresentation in dendritic cells. (a) DC2.4 cells stably transfected with C-terminal mCherry-tagged WT, CA or DN ARF6 by lentivirus were analyzed by live cell imaging. Scale bars are 15 µm. (b) Crosspresentation of soluble OVA by DC2.4 cells in the absence or presence of 80 µM primaquine or 25 µM chloroquine was evaluated by CD69 upregulation in OT-1 cells as described in the section on ‘Methods'. (c) GFP, WT or mutant ARF6-transfected DC2.4 cells were stimulated with soluble OVA at the indicated concentrations for 4 h and then cocultured with OT-1 cells for 24 h. CD69 on OT-1 cells was detected by flow cytometry. (d) qPCR analysis of ARF6 knockdown efficiency in DC2.4 cells transfected with lentivirus encoding ARF6 shRNA. GAPDH was used as internal reference. (e) The same as in c, crosspresentation of soluble OVA by DC2.4 cells transfected with non-target or ARF6 shRNA was evaluated by CD69 upregulation in OT-1 cells. (f) Indels introduced by CRISPR/Cas9 for ARF6 loci in DC2.4 cells. Letters in gray background were sgRNA target sequences. Nucleotide deletions and insertions in five clones are indicated by dashes and underlined letters, respectively. (g) As above, crosspresentation of soluble OVA by vector control DC2.4 cells and five ARF6 knockout clones was evaluated by CD69 upregulation in OT-1 cells. (h and j) Quantification of Rab22a and Rab11a shRNA knockdown in DC2.4 cells. (i and k) Rab22a or Rab11a shRNA-treated DC2.4 cells were analyzed for crosspresentation. Student's t-test results for all figures are indicated as follows: **P<0.01; *P<0.05; P at 0.05 or larger was considered non-significant: ns. DN, dominant-negative; CA, constitutive-active; OVA, ovalbumin; WT, wild-type.