Figure 3.
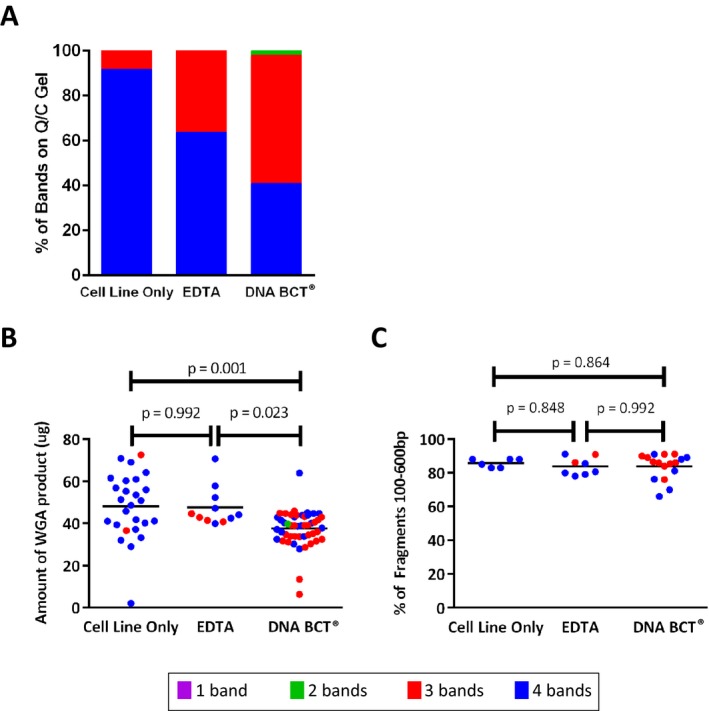
Comparison of whole genome amplification (WGA) product and library prep quality for 3 cell preparation methods. In these experiments, we used MCF7 or HCC1954 cells and the DEPArray to directly generate cell pools of unspiked cells (n = 26), or we spiked tumor cells into normal donor blood. For spiked samples, blood was collected into either EDTA (n = 11) or preservative‐containing DNA BCT ® (n = 49) tubes (86 samples in total, including unspiked). Pools of 10, 20, 50, 100, and 1000 cells were generated. (A) Multiplex PCR of 4 genes was conducted and the indicated numbers of bands detected for each cell preparation method are displayed. (B) Picogreen was used to measure the amount of WGA product for an aliquot of each sample (n = 86), thus ensuring sufficient input material for library preparation and sequencing. (C) Libraries were prepared for 31 of the 86 samples, and the Tapestation‐based percentage of post‐library prep fragments that fall within 100–600 bp was measured. Color of dot for panels B and C indicates number of bands detected by Q/C as depicted in panel A.
