Fig. 3.
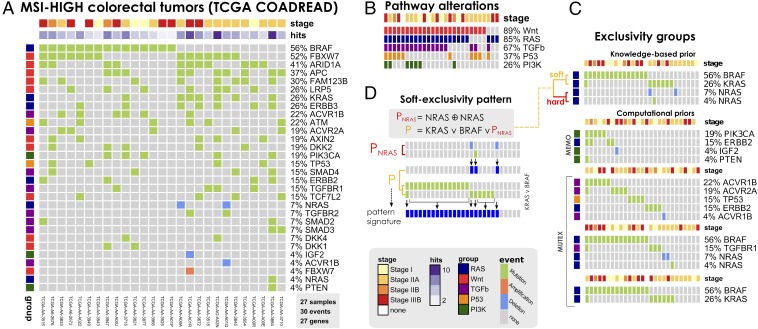
(A) MSI-HIGH colorectal tumors from the TCGA COADREAD project (56), restricted to 27 samples with both somatic mutations and high-resolution CNA data available and a selection out of 33 driver genes annotated to WNT, RAS, PI3K, TGF-β, and P53 pathways. This dataset is used to infer the model in Fig. 5. (B) Mutations and CNAs in MSI-HIGH tumors mapped to pathways confirm heterogeneity even at the pathway level. (C) Groups of mutually exclusive alterations were obtained from ref. 56—which run the MEMO (76) tool—and by MUTEX (77) tool. In addition, previous knowledge about exclusivity among genes in the RAS pathway was exploited. (D) A Boolean formula input to CAPRI tests the hypothesis that alterations in the RAS genes KRAS, NRAS, and BRAF confer equivalent selective advantage. The formula accounts for hard exclusivity of alterations in NRAS mutations and deletions, jointly with soft exclusivity with KRAS and NRAS alterations.