Fig. 3.
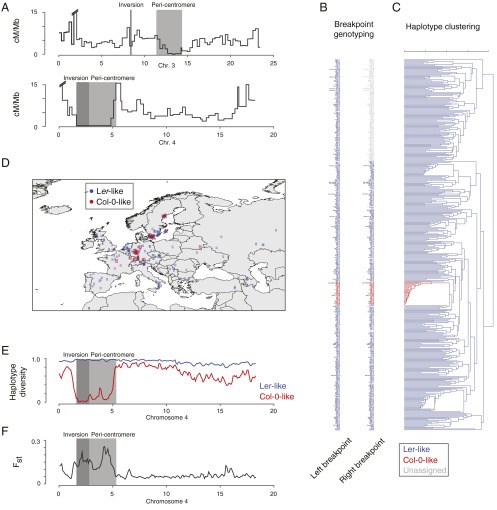
Impact of large-scale inversions on meiotic recombination and haplotype diversity in a worldwide collection of A. thaliana accessions. (A) Male meiotic recombination frequencies across chromosomes 3 and 4 contrasted with the location of the two large-scale inversions (dark gray boxes) and the pericentromeric regions (light gray boxes) [recombination data generated by Giraut et al. (26)]. Recombination frequency was measured between markers with an average distance of 316 kb. Both inversions co-occur with locally reduced recombination frequencies. The interval harboring the inversion on chromosome 3, however, showed residual recombination activity, which does not imply recombination in the inverted region, but might arise from recombination in 111 kb of noninverted sequence in this interval. (B) The names of 409 accessions colored by the inferred chromosome 4 inversion allele (blue, Ler allele; red, Col-0 allele) as assessed on the left and right breakpoints of the inversion. The accessions were ordered after their occurrence in the haplotype clustering shown in C. (C) Haplotype clustering based on 9,198 SNPs located within the chromosome 4 inversion, revealing two distinct clusters, which perfectly matched the two chromosome 4 inversion alleles. (D) Distribution of the accession origins in central Europe, colored by their respective chromosome 4 inversion alleles. (E) Haplotype diversity within the accessions carrying a Col-0–like (red) or Ler-like allele (blue) of the chromosome 4 inversion. (F) Population differentiation (Fst) between these two groups of accessions. Inversion and pericentromere shown with dark and light gray boxes.