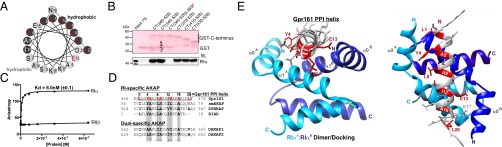
Fig. 2.
Interaction of Gpr161:RIα in vitro and in silico. (A) Helical wheel projection of the helix propensity of Gpr161-CT from L458 to L477 (murine Gpr161 numbering; helix numbering 1–20). Red labeling indicates conserved amino acids potentially involved in PPI with RI. (B) GST pulldown experiments of recombinant RIα in the presence or absence of 5 mM cAMP using indicated GST-CT fusions of Gpr161 including the CT340–528-L465P mutant. (C) Binding analyzes of full-length RIα (●) or full-length RIIβ (■) to the 5-carboxyfluorescein (5-FAM)–labeled peptide of Gpr161 (amino acids 457–481; 25-mer) by steady-state fluorescence anisotropy measurements. ±SD from n = 3 independent experiments. (D) Structure-based alignment of Gpr161 (murine) with RI-specific and dual-specific AKAPs (human). The four binding pockets are highlighted in gray. Conserved and possibly PPI-involved amino acids are shown in bold. Gpr161 residues that seem to participate in the interaction with the hydrophobic groove of the RI-D/D domain are highlighted in red. Identity (*) and similarities (: and .) are indicated. (E) Structural model of RIα D/D domain in complex with Gpr161-CT456-478 peptide. Crystal structure of RIα D/D-domain:DAKAP-2 peptide (PDB ID code 3IM4) was used to build the model. The DAKAP-2 sequence was substituted with the predicted amphipathic helix sequence of Gpr161-CT456–478. Side and top view of the model are shown. The monomers of the RIα D/D domain are depicted in blue and the Gpr161 helix is shown gray with PPI-relevant amino acids highlighted in red (helix numbering 1–20).