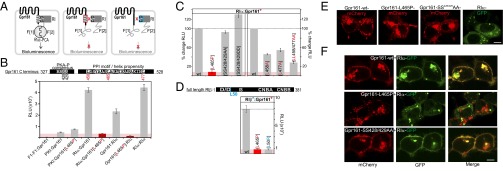
Fig. 3.
Cellular PPIs and localization of Gpr161 variants. (A) Schematic illustration of the Rluc-PCA biosensor strategy to quantify PPIs of wild-type and mutated Gpr161 and RI in vivo. Mutated domains are highlighted in red/blue. (B) Shown are conserved sequence elements in the Gpr161-CT. Impact of L465P mutation of Gpr161-F[1]/[2] on complex formation with RIα-F[1]/[2] (±SEM; representative of n = 3 independent experiments; murine Gpr1611–528; NP_001297359.1). (C) Impact of Gpr161 mutations of the flanking Leu of the PPI-motif and the PKA phosphorylation consensus site on RIα:Gpr161 PPI. Read out: Rluc PCA (±SEM of at least n = 4 independent experiments). (D) Impact of the L50R mutation on RIβ:Gpr161 PPI; Rluc PCA measurements (±SEM; representative of n = 3). (E) Subcellular localization of mCherry-tagged Gpr161 or GFP-tagged RIα hybrid proteins in HEK293 cells. (Scale bar, 5 μm.) (F) Subcellular localization of coexpressed mCherry-tagged Gpr161 variants and GFP-tagged RIα in HEK293 cells. (Scale bar, 5 μm.)